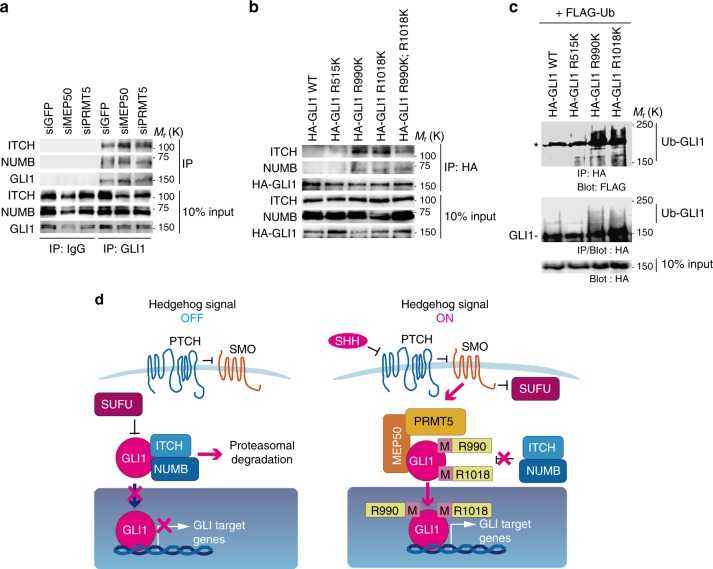
Fig. 5.
MEP50/PRMT5 complex-mediated GLI1 methylation inhibits the interaction of GLI1 with its E3 ligase complex, ITCH/NUMB, resulting in GLI1 stabilisation. a Interaction of GLI1 and endogenous ITCH or NUMB from stably PRMT5-knockdown or MEP50-knockdown C3H10T1/2 cells. siMEP50-m2 and siPRMT5-m2 siRNAs were stably expressed by recombinant retroviruses. MG132 (50 μM) was applied for 4 h before harvesting. b Interaction of GLI1 mutants with endogenous ITCH or NUMB in C3H10T1/2 cells. The cells were transfected as indicated. At 48 h post-transfection, 50 μM MG132 was applied for 4 h, and then the cells were lysed and subjected to immunoprecipitation with an anti-HA antibody, followed by immunoblotting with antibodies against the indicated proteins. c In vivo ubiquitination of HA-GLI1-RK mutants. Cells were transfected and cultured for 24 h, followed by treatment with 50 µM MG132 for 4 h before harvesting. Ubiquitinated GLI1 was detected by immuoprecipitation with an anti-HA (3F10) antibody and immunoblotting with anti-FLAG (upper panel) or anti-HA (lower panel) antibodies. The asterisk denotes non-specific bands. d Schematic diagram of the mechanism of PRMT5/MEP50-mediated GLI1 stabilisation. When the HH signalling pathway inactivates, the ITCH/NUMB E3 ligase complex binds to and ubiquitinates GLI1 for proteasomal degradation. In turn, under HH signalling pathway activation, the MEP50/PRMT5 complex methylates GLI1 to dissociate the ITCH/NUMB complex from GLI1, resulting in GLI1 stabilisation. Unprocessed original scans of blots are shown in Supplementary Fig. 6