Fig. 3.
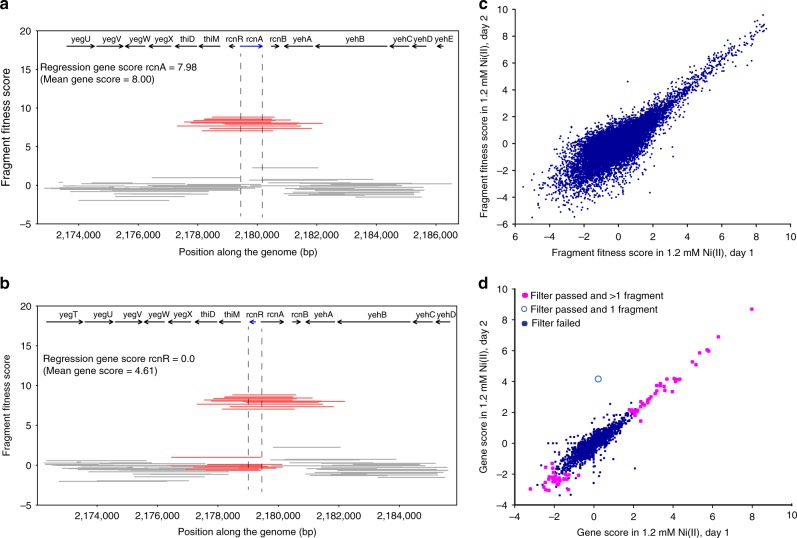
Fragment and gene fitness dual-barcoded shotgun expression library sequencing (Dub-seq) scores. a Dub-seq fragment (strain) data for region surrounding rcnA under elevated nickel stress (y axis). Each line shows a Dub-seq fragment. Those that completely cover rcnA are in red. Both the mean and regression scores reflect the known biology of rcnA as a nickel resistance determinant47. b Same as (a) for the neighboring rcnR, which encodes a transcriptional repressor of rcnA. Fragments that cover rcnR are in red. c Comparison of fragment fitness scores for two biological replicates of 1.2 mM nickel stress. d Same as (c) for gene fitness scores calculated using the regression approach. Genes are highlighted if their score met our criteria for a high-confidence effect within an experiment (without considering other experiments) (see Methods); we also show whether the gene score is based on just one fragment (effects from just one fragment are not considered high confidence if they do not replicate). Source data are provided as a Source Data file