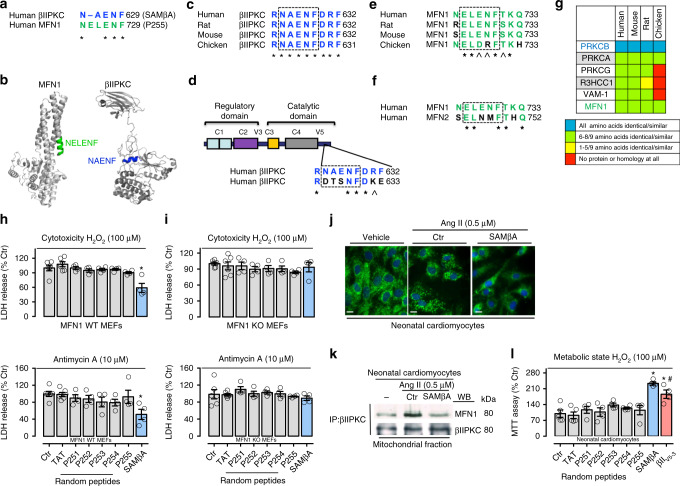
Fig. 5.
Rational design of peptide that selectively inhibits Mfn1 and βIIPKC interaction. a Sequence alignment of human βIIPKC and Mfn1 identified a short sequence of homology, RNAENF/NELENF. b NELENF in Mfn1 (PDB: 5GOM) and RNAENF in the V5 domain of βIIPKC (PDB: 3PFQ) are exposed and available for protein−protein interactions (see colored structures). Note that the alternatively spliced βIPKC varies from βIIPKC only in the V5 domain15. c Conservation of RNAENFDRF sequence in βIIPKC in a variety of species. d RNAENFDRF sequence is not present in the V5 domain of βIPKC, a βIIPKC alternative spliced form. e Conservation of NELENFTKQ in Mfn1 in a variety of species. f NELENFTKQ sequence is not present in Mfn2. g RNAENFDRF sequence is found in 6 human proteins. Heat-map of the RNAENFDRF conservation in orthologous of these proteins shows RNAENFDRF conservation only in βIIPKC. (Further information about all the proteins is given in the Supplementary Table 2). *Denotes identity, and ^ denotes homology. h Mfn1 wild type (WT, n = 4–6) and i Mfn1 knockout (KO, n = 4–6) MEFs were treated with TAT, p251–255 or SAMβA peptides for 30 min followed by incubation with either H2O2 (100 μM, 24 h) or Antimycin A (10 μM, 24 h). After incubation, cytotoxicity was measured by LDH release. j Neonatal rat cardiomyocytes in culture were treated with TAT or SAMβA for 30 min followed by incubation with angiotensin II (0.5 µM for 4 h, n = 5 per group). Cells were then stained with anti-Tom20 antibody (green) and Hoechst stain and counted. Mitochondrial morphology was analyzed using a ×63 oil immersion lens (scale bar: 10 μm). k Co-immunoprecipitates with anti-βIIPKC were analyzed in the mitochondrial fraction by western blot (representative blots of three independent experiments). l Neonatal rat cardiomyocytes in culture were treated with TAT, P251–255, SAMβA or βIIV5-3 peptides for 30 min followed by incubation with H2O2 (100 μM for 24 h, n = 4–5 per group). After incubation, cell viability was measured by MTT assay. Data are means ± SEM. *P < 0.05 vs. Ctr. #P < 0.05 vs. SAMβA-treated cells. One-way analysis of variance (ANOVA) with post-hoc testing by Duncan