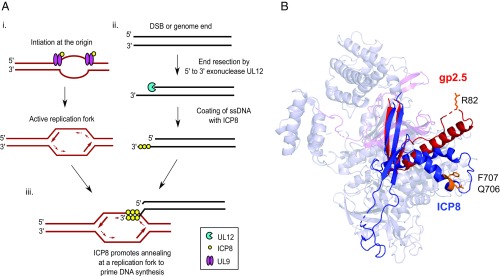
Fig. 8.
Role of ICP8 annealing activity during HSV-1 DNA replication. (A) A model for HSV-1 RDR. (i) ICP8 and UL9 distort and unwind the origin to create an active replication fork. (ii) A free genome end or double-strand break created during infection is resected by a 5′-to-3′ exonuclease and the resulting 3′ overhang coated by ICP8. (iii) ICP8 promotes annealing of the resulting 3′-ssDNA overhang to an active replication fork to prime DNA synthesis. (B) Structural alignment of T7 bacteriophage SSAP gp2.5 (red; PDB ID code 1JE5; chain A) and ICP8 (blue; PDB ID code 1URJ; chain A) using jCE circular permutation algorithm. ICP8 Q706/F707 residues and gp2.5 R82 residue are illustrated in orange.