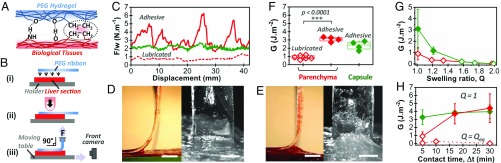
Fig. 1.
Ex vivo adhesion on freshly dissected liver tissues. (A) Schematic representation of the hydrogen-bonding and hydrophobic interactions between PEG hydrogels and biological tissues. (B) Schematic representation of the peeling protocol. (C) Normalized peeling force as a function of displacement on the liver capsule (green curve) and parenchyma (red curves) for PEG ribbons (Q = 1, Δt = 5 min). The origin of displacement (0 mm) corresponds to the beginning of the steady-state peeling. (D and E) Side and front views during peeling of a PEG ribbon (Q = 1, Δt = 5 min) from liver parenchyma displaying (D) a lubricated regime or (E) an adhesive regime. (Scale bar: 5 mm.) (F) Adhesion energy for peeling of PEG ribbons (Q = 1, Δt = 5 min) on liver capsule (green) and parenchyma (red). In this graph and the following, full and open symbols correspond to adhesive and lubricated behaviors, respectively. (G) Adhesion energy as a function of hydrogel swelling ratio for Δt = 5 min: liver capsule (green) and parenchyma (red). (H) Adhesion energy as a function of contact time for dry PEG ribbons (Q = 1): liver capsule (green) and parenchyma (red). Dotted line shows data for fully swollen PEG ribbons (Q = Qeq).