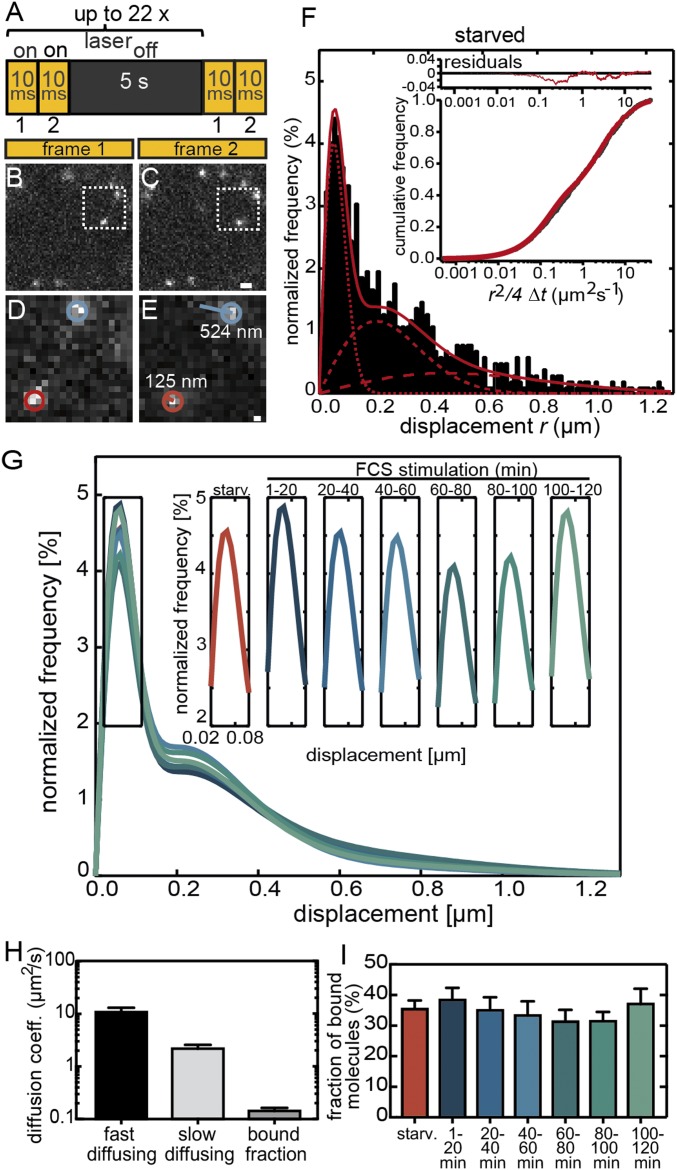
Fig. 2.
Analysis of Halo-SRF binding fractions in unstimulated and stimulated fibroblasts. (A) Illumination scheme indicating on and off times of laser illumination. (B and C) Typical jump distances of individual molecules between two frames are illustrated by two exemplary molecules. (D and E) Magnified areas of the regions marked by a dashed boxes in B and C. The molecule labeled in blue was mobile and moved by 524 nm, whereas the red molecule was rather immobile and had a jump distance of only 125 nm. (Scale bar: B and C, 1 μm; D and E, 160 nm.) (F) Distribution of single-molecule displacements in 10-ms integration time (n = 1,018 molecules, n = 21 cells) normalized to all molecules detected in serum-starved NIH 3T3 cells (black bars). The distributions were fitted with a three-component diffusion model (red line; single components in dotted red lines; SI Appendix, Eqs. I and II) where the lowest jump distances are representative of chromatin-bound molecules. (Inset) The residuals (red line in upper part) from the three-component fit (red line in lower part) to the cumulative distribution of all calculated squared displacements per 10-ms integration time for a 2D diffusion (in black). (G) Three-component fits including three diffusion coefficients to the measured displacement histograms for starved NIH 3T3 cells and consecutive 20-min time intervals of serum stimulation (for N numbers, see SI Appendix, Fig. S1). (Inset) Changes in the amplitude of the bound fraction only for all 20-min time intervals. (H) Average diffusion coefficients (mean ± SEM) for all three Halo-SRF fractions determined from a global fit to cumulative histograms of unstimulated and stimulated cells computed from all timepoints of serum stimulation. Values were calculated by bootstrapping. (I) The ratio of bound molecules to all molecules is depicted in starved cells and along the several timepoints of FCS application (mean ± SEM). Between 1 min to 20 min and 100 min to 120 min, FCS slightly enhanced the fraction of bound molecules. Values were calculated by bootstrapping.