Abstract
The critical state of intensive care unit (ICU) patients demands close monitoring, and as a result a large volume of multi-parameter data is collected continuously. This represents a unique opportunity for researchers interested in clinical data mining. We sought to foster a more transparent and efficient intensive care research community by building a publicly available ICU database, namely Multiparameter Intelligent Monitoring in Intensive Care II (MIMIC-II). The data harnessed in MIMIC-II were collected from the ICUs of Beth Israel Deaconess Medical Center from 2001 to 2008 and represent 26,870 adult hospital admissions (version 2.6). MIMIC-II consists of two major components: clinical data and physiological waveforms. The clinical data, which include patient demographics, intravenous medication drip rates, and laboratory test results, were organized into a relational database. The physiological waveforms, including 125 Hz signals recorded at bedside and corresponding vital signs, were stored in an open-source format. MIMIC-II data were also deidentified in order to remove protected health information. Any interested researcher can gain access to MIMIC-II free of charge after signing a data use agreement and completing human subjects training. MIMIC-II can support a wide variety of research studies, ranging from the development of clinical decision support algorithms to retrospective clinical studies. We anticipate that MIMIC-II will be an invaluable resource for intensive care research by stimulating fair comparisons among different studies.
I. INTRODUCTION
Critically ill patients in intensive care units (ICU) are closely monitored, resulting in extensive collections of detailed physiologic, biochemical, imaging, pharmacologic, and clinical data. The abundant data supports the diagnostic and management efforts of the clinicians, even though some have complained of “data overload”. It has become technically feasible to collect and archive this rich stream of data, and to create a unique ICU database capable of supporting a wide variety of retrospective critical care research, and the development and validation of automated clinical decision support algorithms. Making such a database freely available to the research community will significantly enhance the rate of such research.
In this paper we present the development and characteristics of the Multiparameter Intelligent Monitoring in Intensive Care II (MIMIC-II) database (The predecessor of MIMIC-II, namely MIMIC, is described in [1]). We first describe the data collection, followed by details of the database structure and deidentification process. Subsequently, we present patient statistics of MIMIC-II (version 2.6) and discuss the kinds of research that can be conducted using MIMIC-II. For a more complete but slightly older description of MIMIC-II (version 2.4), including comparisons with other ICU databases, please see [2].
II. METHODS
A. Data Collection
The ICU data in MIMIC-II were collected at Beth Israel Deaconess Medical Center (BIDMC) in Boston, MA, USA during the period from 2001 to 2008. Adult data were acquired from four ICUs at BIDMC: medical (MICU), surgical (SICU), coronary care unit (CCU), and cardiac surgery recovery unit (CSRU). MIMIC-II also contains data from the neonatal ICU (NICU) of BIDMC, but this paper focuses only on the adult data, which make up the majority of MIMIC-II. This study was approved by the Institutional Review Boards of BIDMC and the Massachusetts Institute of Technology.
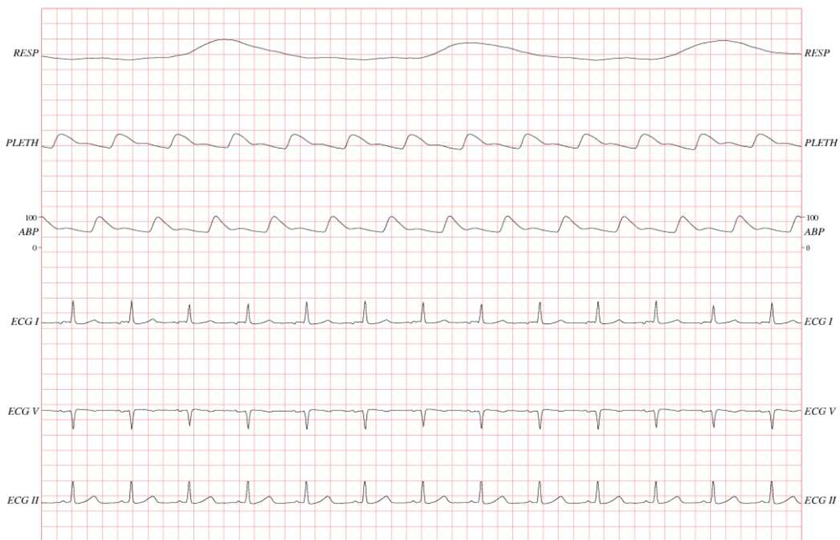
Two types of data were obtained: clinical data and physiological waveforms. The clinical data were acquired from the CareVue Clinical Information System (models M2331A and M1215A; Philips Healthcare, Andover, MA) and the hospital’s electronic archives. The data included patient demographics, nursing notes, discharge summaries, continuous intravenous drip medications, laboratory test results, nurse-verified hourly vital signs, etc. Table I describes different clinical data types in MIMIC-II by giving examples of each type. The physiological waveforms were collected from bedside monitors (Component Monitoring System Intellivue MP-70; Philips Healthcare) and included high-resolution (125 Hz) waveforms (e.g., electrocardiograms), derived time series such as heart rate, blood pressures, and oxygen saturation (either once-per-minute or once-per-second), and monitor-generated alarms. Figure 1 shows an example of high-resolution waveforms.
TABLE I.
Clinical data types in MIMIC-II
| Clinical Data Type | Examples |
|---|---|
| Demographics | Age, gender, date of death (date-shifted, in-hospital or after discharge), ethnicity, religion |
| Hospital admission | Admission and discharge dates (date-shifted), room tracking, code status, ICD-9 codes, DRG |
| Intervention | Ventilator settings, IV medications, provider order entry data, CPT codes |
| Laboratory tests | Blood chemistries, hematology, urinalysis, microbiologies |
| Fluid balance | Solutions, blood transfusion, urine output, estimated blood loss |
| Free-text | Reports of imaging studies (no actual images) and 12-lead ECGs, nursing notes, hospital discharge summaries |
| Severity scores | SAPS I, SOFA, Elixhauser comorbidities |
ICD-9, International Classification of Diseases, 9th Revision; DRG, Diagnosis Related Group; IV, intravenous; ECGs, electrocardiograms; CPT, Current Procedural Terminology; SAPS, Simplified Acute Physiological Score; SOFA, Sequential Organ Failure Assessment
Fig. 1.
An example of high-resolution waveforms
B. Database Organization
After data collection, the clinical data were processed and imported into a relational database that can be queried using Structured Query Language [3]. The database was organized according to individual patients at the highest level. A given patient might have had multiple hospital admissions and each hospital admission in turn could have included multiple ICU stays; within the same hospital admission, ICU stays separated by a gap greater than 24 hours were counted separately. Unique subject, hospital admission, and ICU stay IDs were linked to one another to indicate relationships among patients, hospital admissions, and ICU stays.
The physiological waveforms were converted from the proprietary Philips format to an open source format (WFDB)[4] to be stored separately from the clinical data. Because the clinical and physiological data originated from different sources, they had to be matched to each other by confirming a common patient source [5]. Although unique identifiers such as medical record number and patient name were utilized for this matching task, a significant portion of the physiological waveforms lacked such an identifier, resulting in limited matching success. Moreover, waveform data collection spanned a shorter period of time than clinical data collection due to technical issues, and waveform data were not collected in the first place for many ICU stays.
C. Deidentification
In order to comply with Health Insurance Portability and Accountability Act, MIMIC-II was deidentified by removing protected health information (PHI). Also, the entire time course of each patient (all hospital admissions and ICU stays) was time-shifted to a hypothetical period in the future. This deidentification was a straight-forward task for structured data fields but was a challenging task for free-text data such as nursing notes and discharge summaries. Thus, an automated deidentification algorithm was developed and was shown to perform better than human clinicians in detecting PHI in free-text documents. For more details about this open-source algorithm, please see [6], [7].
D. Public Access
In order to gain free access to MIMIC-II, any interested researcher simply needs to complete a data use agreement and human subjects training. The actual access occurs over the Internet. The clinical data can be accessed either by downloading a flat-file text version or via a live connection through password-protected web service. The physiological waveforms are best accessed using the WFDB software package. For detailed information regarding obtaining access to MIMIC-II, please see the MIMIC-II website: http://physionet.org/mimic2.
III. RESULTS
Table II tabulates adult patient statistics in MIMIC-II, stratified with respect to the four critical care units. In total, 26,870 adult hospital admissions and 31,782 adult ICU stays were included in MIMIC-II. MICU patients formed the largest proportion among the 4 care units, while CCU patients made up the smallest cohort. Only 15.7% of all ICU stays were successfully matched with waveforms. In terms of neonates, 7,547 hospital admissions and 8,087 NICU stays were added to MIMIC-II.
TABLE II.
Adult patient statistics in MIMIC-II (version 2.6), stratified with respect to critical care unit
| MICU | SICU | CSRU | CCU | Total | |
|---|---|---|---|---|---|
| Hospital admissions1 | 10,313 (38.4%) | 6,925 (25.8%) | 5,691 (21.2%) | 3,941 (14.7%) | 26,870 (100%) |
| Distinct ICU stays2 | 12,648 (39.8%) | 8,141 (25.6%) | 6,367 (20.0%) | 4,626 (14.6%) | 31,782 (100%) |
| Matched waveforms3 | 2,313 (18.3%) | 673 (8.3%) | 1,195 (18.8%) | 798 (17.3%) | 4,979 (15.7%) |
| Age (yrs)4 | 64.5 (50.1, 78.2) | 61.1 (46.7, 75.9) | 67.1 (57.0, 76.2) | 71.4 (58.9, 80.7) | 65.5 (51.9, 77.7) |
| Gender (male)3 | 6,301 (49.8%) | 4,701 (57.7%) | 4,147 (65.1%) | 2,708 (58.5%) | 17,857 (56.2%) |
| ICU length of stay (days)4 | 2.1 (1.1, 4.3) | 2.4 (1.2, 5.4) | 2.1 (1.1, 4.1) | 1.9 (1.0, 3.5) | 2.1 (1.1, 4.3) |
| Hospital length of stay (days)4 | 7 (4, 13) | 8 (5, 16) | 8 (5, 12) | 5 (3, 9) | 7 (4, 13) |
| First day SAPS I4 | 13 (10, 17) | 14 (10, 17) | 17 (14, 20) | 12 (9, 15) | 14 (10, 18) |
| Mechanical ventilation3 | 4,202 (33.2%) | 4,131 (50.7%) | 5,152 (80.9%) | 1,076 (23.3%) | 14,561 (45.8%) |
| Swan-Ganz hemodynamic monitoring3 | 366 (2.9%) | 1,066 (13.1%) | 4,137 (65.0%) | 1,086 (23.5%) | 6,655 (20.9%) |
| Invasive arterial blood pressure monitoring3 | 3,944 (31.2%) | 5,343 (65.6%) | 5,545 (87.1%) | 2,054 (44.4%) | 16,886 (53.1%) |
| Use of vasoactive medications3 | 2,859 (22.6%) | 1,982 (24.4%) | 4,397 (69.1%) | 1,334 (28.8%) | 10,572 (33.3%) |
| Hospital mortality3 | 1,645 (16%) | 842 (12.2%) | 213 (3.7%) | 392 (10.0%) | 3,092 (11.5%) |
This table is an updated version of Table in [2], which is based on version 2.4.
MICU, medical ICU; SICU, surgical ICU; CSRU, cardiac surgery recovery unit; CCU, coronary care unit; SAPS, Simplified Acute Physiological Score
N (% of total admissions)
N (% of total ICU stays)
N (% of unit stays)
median (first quartile, third quartile)
Among the adults, the overall median ICU and hospital lengths of stay were 2.1 and 7 days, respectively. CSRU patients were characterized by high utilization of mechanical ventilation, Swan-Ganz, invasive arterial blood pressure monitoring, and vasoactive medications. Overall, 45.8% and53.1% of all adult ICU stays utilized mechanical ventilation and invasive arterial blood pressure monitoring, respectively. In-hospital mortality rate was highest in the MICU (16%) and lowest in the CSRU (3.7%). The overall in-hospital mortality was 11.5%.
IV. DISCUSSION
In MIMIC-II, we have successfully created a publicly available database for the intensive care research community. MIMIC-II is a valuable resource, especially for those researchers who do not have easy access to the clinical intensive care environment. Furthermore, research studies based on MIMIC-II can be compared with one another in an objective manner, which would reduce redundancy in research and foster more streamlined advancement in the research community as a whole.
The diversity of data types in MIMIC-II opens doors for a variety of research studies. One important type of research that can stem from MIMIC-II is the development and evaluation of automated detection, prediction, and estimation algorithms. The high temporal resolution and multi-parameter nature of MIMIC II data are suitable for developing clinically useful and robust algorithms. Also, it is easy to simulate a real-life ICU in offline mode, which enables inexpensive evaluation of developed algorithms without the risk of disturbing clinical staff. Previous MIMIC-II studies in this research category include hypotensive episode prediction[8] and robust heart rate and blood pressure estimation [9]. Additional signal processing studies based on MIMIC-II include false arrhythmia alarm suppression [10] and signal quality estimation for the electrocardiogram [11].
Another type of research that MIMIC-II can support is retrospective clinical studies. While prospective clinical studies are expensive to design and perform, retrospective studies are inexpensive, demand substantially less time-commitment, and allow flexibility in study design. MIMIC-II offers severity scores such as the Simplified Acute Physiological Score I [12] and Sequential Organ Failure Assessment [13] that can be employed in multivariate regression models to adjust for differences in patient conditions. For example, Jia and colleagues [14] investigated risk factors for acute respiratory distress syndrome in mechanically ventilated patients, and Lehman and colleagues [15] studied hypotension as a risk factor for acute kidney injury.
MIMIC-II users should note that real-life human errors and noise are preserved in MIMIC-II since no artificial cleaning or filtering was applied. Although this presents a challenge, it also is an opportunity for researchers to work with real data and address pragmatic issues.
Because MIMIC-II is a single-center database originating from a tertiary teaching hospital, research results stemming from MIMIC-II may be subject to institutional or regional bias. However, many research questions can be answered independent of local culture or geographic location (e.g., the focus of the study is physiology).
A successful MIMIC-II study requires a variety of expertise. While clinically-relevant research questions would best come from clinicians, reasonable database and computer skills are necessary to extract data from MIMIC-II. Hence, a multi-disciplinary team of computer scientists, biomedical engineers, biostatisticians, and intensive care clinicians is strongly encouraged in designing and conducting a research study using MIMIC-II.
V. CONCLUSIONS
MIMIC-II is a large ICU database that encompasses detailed patient demographics, records of clinical interventions, physiological waveforms and vital signs, and much more. Its public availability contributes to building a vigorous and collaborative research community.
VI. ACKNOWLEDGMENTS
This work was supported by grant R01 EB001659 from the National Institute of Biomedical Imaging and Bioengineering, and by the Massachusetts Institute of Technology, Beth Israel Deaconess Medical Center, and Philips Healthcare. The authors of this article would like to acknowledge the following people for their contributions to the development of MIMIC-II: Tin H. Kyaw, Benjamin E. Moody, GeorgeB. Moody, Ishna Neamatullah, Margaret Douglass, WilliamH. Long, Li-Wei H. Lehman, Xiaoming Jia, Michael Craig, Larry Nielsen, Greg Raber, and Brian Gross.
REFERENCES
- [1].Moody GB and Mark RG, “A database to support development and evaluation of intelligent intensive care monitoring,” Computers in Cardiology, vol. 33, pp. 657–660, 1996. [Google Scholar]
- [2].Saeed M, Villarroel M, Reisner AT, Clifford GD, Lehman L,Moody G, Heldt T, Kyaw TH, Moody B, and Mark RG, “Multiparameter Intelligent Monitoring in Intensive Care II (MIMIC-II): a public-access intensive care unit database,” Critical Care Medicine, vol. 39, no. 5, pp. 952–960, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Price J, Oracle Database 11g SQL. McGraw-Hill Osborne Media, 2007. [Google Scholar]
- [4].The WFDB software package. [Online]. Available: www.physionet.org/physiotools/wfdb.shtml
- [5].Craig M, Moody B, Jia S, Villarroel M, and Mark R, “Matching data fragments with imperfect identifiers from disparate sources,” Computing in Cardiology, vol. 37, pp. 793–796, 2010. [Google Scholar]
- [6].De-identification: software and test data. [Online]. Available: www.physionet.org/physiotools/deid/
- [7].Neamatullah I, Douglass MM, Lehman LH, Reisner A, Villarroel M, Long WJ, Szolovits P, Moody B, Mark RG, and Clifford GD, “Automated de-identification of free-text medical records,” BMC Medical Informatics and Decision Making, vol. 8:32, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Lee J and Mark RG, “An investigation of patterns in hemodynamic data indicative of impending hypotension in intensive care,” BioMedical Engineering Online, vol. 9:62, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Li Q, Mark RG, and Clifford GD, “Artificial arterial blood pressure artifact models and an evaluation of a robust blood pressure and heart rate estimator,” BioMedical Engineering Online, vol. 8:13, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Aboukhalil A, Nielsen L, Saeed M, Mark RG, and Clifford GD, “Reducing false alarm rates for critical arrhythmias using the arterial blood pressure waveform,” Journal of Biomedical Informatics, vol. 41, no. 3, pp. 442–451, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Li Q, Mark RG, and Clifford GD, “Robust heart rate estimation from multiple asynchronous noisy sources using signal quality indices and a Kalman filter,” Physiological Measurement, vol. 29, pp. 15–32, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Le Gall JR, Loirat P, and Alperovitch A, “Simplified acute physiological score for intensive care patients,” Lancet, vol. 322, no. 8352, p. 741, 1983. [DOI] [PubMed] [Google Scholar]
- [13].Vincent J, Moreno R, Takala J, Willatts S, De Mendonca A,Bruining H, Reinhart C, Suter P, and Thijs L, “The SOFA (Sepsis-related Organ Failure Assessment) score to describe organ dysfunction/failure,” Intensive Care Medicine, vol. 22, no. 7, pp. 707–710, 1996. [DOI] [PubMed] [Google Scholar]
- [14].Jia X, Malhotra A, Saeed M, Mark RG, and Talmor D, “Risk factors for ARDS in patients receiving mechanical ventilation for > 48 h,” Chest, vol. 133, no. 4, pp. 853–861, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Lehman L, Saeed M, Moody G, and Mark R, “Hypotension as a risk factor for acute kidney injury in ICU patients,” Computing in Cardiology, vol. 37, pp. 1095–1098, 2010. [PMC free article] [PubMed] [Google Scholar]