Figure 5.
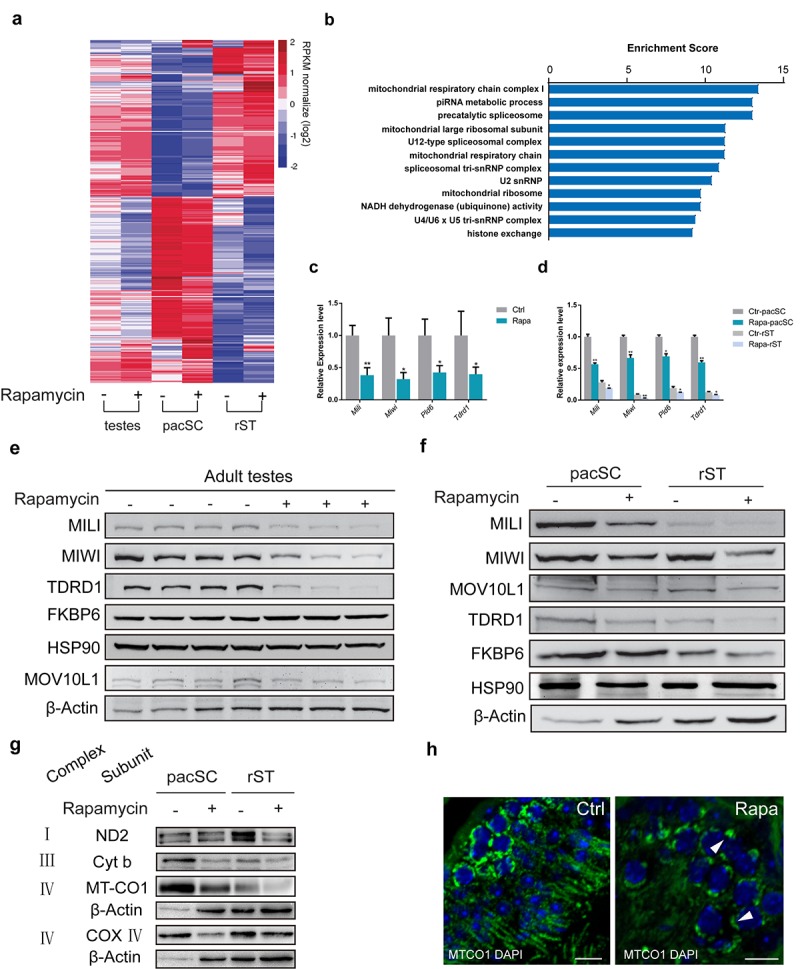
Chronic rapamycin treatment affects the expression of mitochondrial and piRNA pathway genes. Samples were testis tissue (3 control, 3 rapamycin-treated males) and pachytene spermatocytes (pacSC) and round spermatids (rST) isolated from pooled samples (control, n=7 males; rapamycin, n=15). (a) Heatmap of differentially expressed genes. (b) Gene ontology analysis of genes that were downregulated in pachytene spermatocytes from rapamycin-treated mice versus control. (c) Quantitative RT-PCR analysis of piRNA pathway component transcripts in testis tissue. Error bars represent SD (*P < 0.05, Student’s t test). (d) Relative mRNA level of piRNA pathway genes in pachytene spermatocytes and round spermatid populations. Error bars represent SD (*P < 0.05, Student’s t test). (e, f) Western blot analysis of piRNA pathway associated proteins in testis (e), and isolated spermatocytes and round spermatids (f) from control (-) and rapamycin-treated (+) males. (g) Western blot analysis of OXPHOS subunits. β-Actin served as loading control. (h) Immunostaining of testis sections for MT-CO1 (green). Arrowheads mark accumulation of MT-CO1 at the periphery of nuclei in rapamycin-treated testis. Scale bar, 20μm.
