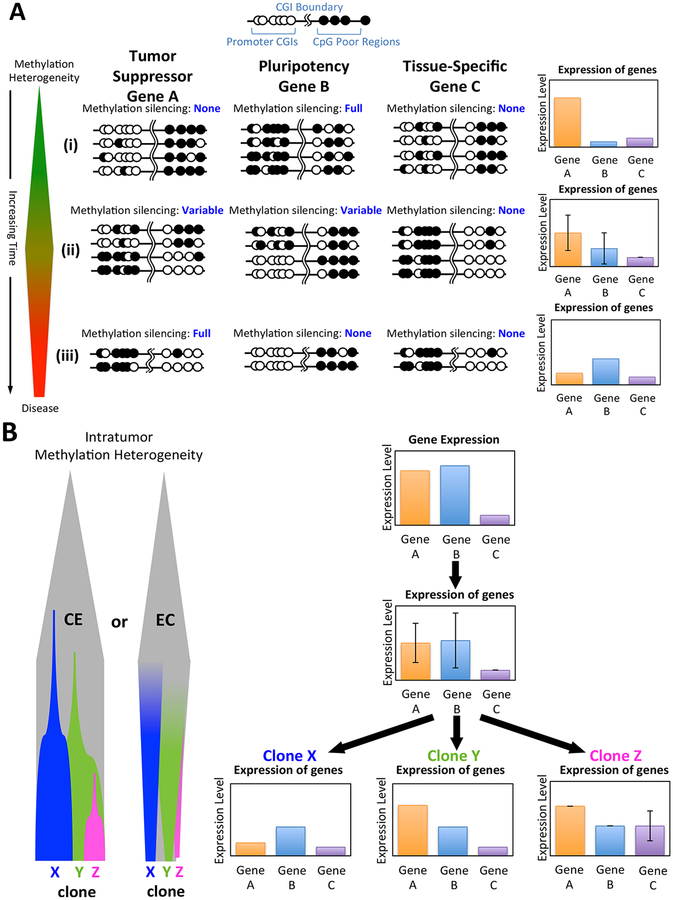
Figure 2. The contribution of heterogeneous methylation to the emergence of cancerous clones.
(A) At the earliest stage (i), minor stochastic differences in methylation patterns accumulate in the promoters of Tumor Suppressor Gene A, Pluripotency Gene B and Tissue-Specific Gene C. Immediately prior to the development of neoplasia (ii), significant methylation heterogeneity has developed in Gene A, resulting in considerable variance in expression between cells. Some select cells become hypomethylated in Gene B, resulting in a pluripotent phenotype. Gene C is not normally expressed and preferentially accrues high levels of CGI methylation in its promoter. As the cancer develops (iii), methylation heterogeneity is somewhat reduced as quasi stable clone X emerges via widespread methylation-mediated silencing of Gene A and hypomethylation of Gene B. Gene C remains unexpressed with heavy methylation in its promoter. (B) The phenotype of Clone X might emerge from methylation/epigenetic heterogeneity through clonal expansion (CE) of a cell that develops a particular phenotypic advantage or through evolutionary convergence (EC), where shared evolutionary pressures yield groups of cells with similar methylation profiles. These same phenomena can yield other phenotypic clones (Clones Y and Z) throughout the course of carcinogenesis, contributing to overall intratumor heterogeneity.