Abstract
Serine integrases catalyze the site-specific insertion of viral DNA into a host’s genome. The minimal requirements and irreversible nature of this integration reaction has led to the use of serine integrases in applications ranging from bacterial memory storage devices to gene therapy. Our understanding of how the integrase proteins recognize the viral (attP) and host (attB) attachment sites is limited, with structural data available for only a Listeria integrase C-terminal domain (CTD) bound to an attP half-site. Here we report quantitative binding and saturation mutagenesis analyses for the Listeria innocua prophage attP site and a new 2.8 Å crystal structure of the CTD•attP half site. We find that Int binds with high affinity to attP (6.9 nM), but the Int CTD binds to attP half-sites with only 7–10 fold lower affinity, supporting the idea that free energy is expended to open an Int dimer for attP binding. Despite the 50 base pair Int-attP interaction surface, only 20 residues are sensitive to mutagenesis and of these, only 6 require a specific residue for efficient Int binding and integration activity. One of the integrase DNA-binding domains, the recombinase domain, appears to be primarily non-specific. Several substitutions result in an improved attP site, indicating that higher efficiency attachment sites can be obtained through site engineering. These findings advance our understanding of serine integrase function and provide important data for efforts towards engineering this family of enzymes for a variety of biotechnology applications.
Keywords: phage integrase, serine integrase, attachment site, integration, specificity
Introduction
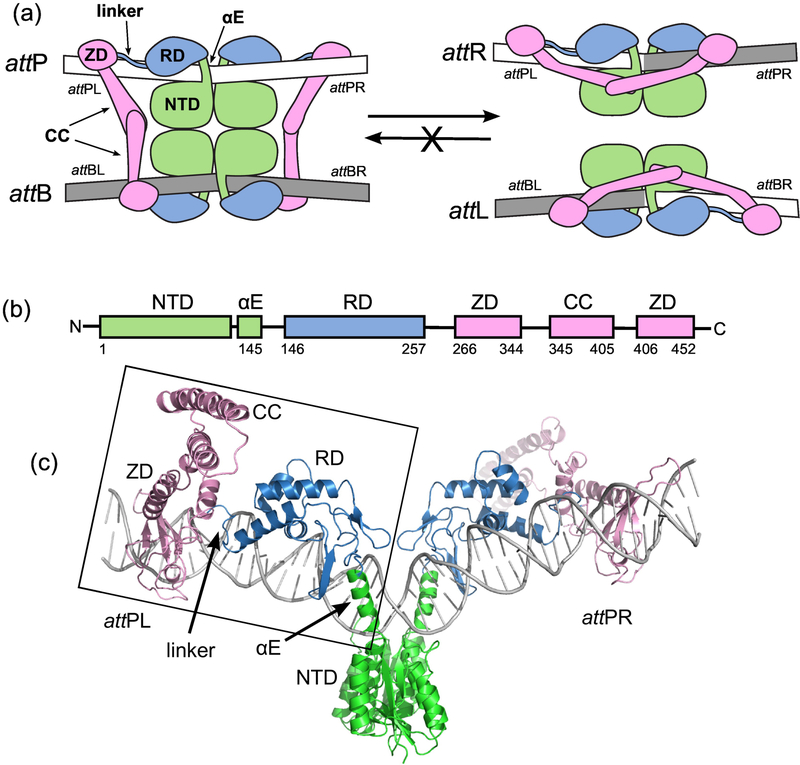
Serine integrases are bacteriophage enzymes that bind to the DNA attachment sites attP and attB, bring the sites together using protein-protein interactions, and carry out site-specific recombination (SSR) to generate two new sites, attL and attR (Fig. 1; [1]). This reaction is used to integrate the phage genome (which contains attP) into that of a host bacterium (which contains attB) to begin a lysogenic life cycle for the virus. Serine integrases are members of the serine recombinase superfamily; these enzymes share a conserved catalytic domain which uses a serine nucleophile to cleave DNA substrates at the start of the SSR reaction [2]. In addition to the conserved catalytic domain, the serine integrases have large carboxyterminal domains (CTDs) that bind to DNA and make inter-domain interactions that play key regulatory roles (Fig. 1). The other large family of SSR enzymes, the tyrosine recombinases, use a tyrosine nucleophile and an entirely different mechanism to accomplish strand exchange between the recombining sites [3]. The mechanistic details of the biochemistry carried out by these enzyme families are reviewed in [2,4,5].
Figure 1.
The integration reaction carried out by serine integrases. A) Schematic of the overall reaction. Integrase dimers bind to attP and attB and associate the sites to form the complex shown on the left. Site-specific recombination catalyzed by the N-terminal catalytic domains (NTDs) results in the hybrid sites attL and attR (right). The reverse reaction that converts attL and attR to attP and attB does not occur at a measurable rate in the absence of a phage encoded RDF protein. For detailed descriptions of the catalytic mechanism, see [2,5]. attPL and attPR refer to the left and right half-sites of attP. B) Domain structure of LI Int. αE is an alpha helix that extends from the NTD to the recombinase domain (RD). A coiled-coil (CC) motif is embedded in the zinc ribbon domain (ZD). C) Structural model of the Int-attP complex. The boxed region corresponds to the experimental LI CTD•attPL crystal structure and has been duplicated to model the full site [25]. The nature of the bend at the center of the site is not currently known.
Several features of the integration reaction shown in Fig. 1 have generated considerable interest from a broad range of scientists, leading to the development of new tools, applications, and devices that make use of serine integrases [6–8]. The first feature is the simplicity of the system. The attP and attB sites are short, generally 40–50 bp in length. The reaction requires only the integrase protein, and the DNA substrates have no strict topological requirements [9,10]. The serine integrase reaction is therefore quite different than integration catalyzed by the phage λ integrase (a tyrosine recombinase), which uses a 240 bp supercoiled attP site and requires the E. coli integration host factor to stabilize sharp DNA bends [11]. The serine integrase reaction is efficient in vitro and in eukaryotic cells [12,13], a property shared by the widely used Cre recombinase [3,14]. Perhaps the most important and intriguing feature is that the integration reaction is effectively irreversible; the attL and attR sites are not recombined by the integrase to regenerate attP and attB, leading to exceptionally stable integration of DNA elements [15]. For these reasons, serine integrases have been adapted for use in a number of technologies, including genome engineering [16], gene therapy [17], synthetic biology [6], and creation of logic circuits and data storage in living cells [18,19].
In synthetic biology applications, having access to an array of independent recombination sites is an important goal. In genomic applications, a major goal is the ability to perform targeted integration at specific sequences. For both examples, as well as many other applications, understanding the sequence requirements for the sites in some detail would facilitate expanding the available sites through directed evolution [20] and/or rational design that exploits the modular nature of the protein-DNA interfaces. Currently, there is only limited understanding of attachment site sequence specificity for a small number of systems. Some studies have used genomic pseudo-attP or pseudo-attB sites to infer the sequence dependence of recombination [21,22] and others have examined the properties of attachment site mutants [23,24].
We recently described the structure of a serine integrase CTD bound to an attP half-site using X-ray crystallography [25]. We used the integrase from a Listeria innocua prophage (LI Int) for this work based on its favorable biochemical properties and our ability to obtain diffracting crystals of protein-DNA complexes. The structure revealed four regions in the CTD that contact the attP site: the αE helix that connects the catalytic domain to a recombinase domain (RD), the RD, the linker connecting the RD to a zinc ribbon domain (ZD), and the ZD (Fig. 1). Several key protein-DNA contacts identified in the structure were tested by single substitutions of the attP sequence, thereby identifying some of the elements in the attP site that are important for integrase binding and recombination. However, there are a relatively small number of direct protein-DNA contacts observed in the crystal structure, leaving most of the attP sequence unexplored. The LI Int system has also led to high resolution structural models that explain how the coiled-coil domains of the integrase interact [26]; these interdomain contacts are thought to regulate the directionality of recombination [25,27]. The closely related Listeria phage A118 integrase has also been characterized [28] and the recombination directionality factor (RDF) responsible for allowing prophage excision has been studied [29]. Thus, the Listeria integrases have become important model systems for understanding serine integrase structure and function.
Here we report the DNA-binding properties and sequence requirements of the Listeria innocua attP site. We find that Int binds to attP with high affinity, with Kd=6.9±0.7 nM. Int also binds to the attP half-sites with high affinity, supporting the idea that free energy is expended upon opening the Int dimer to bind to the attachment site. To obtain a broader understanding of the role of individual attP residues, we have analyzed all possible symmetric substitutions at each position for their ability to undergo in vitro integration, in vivo integration, and Int binding. Although the 50 base pair length of the LI attP sequence implies a high degree of specificity, the majority of positions within the site can be substituted with no consequence on the efficiency of integration. In several cases, integration efficiency increases upon substitution. We also report a new crystal structure of the LI Int CTD•attP half-site, determined at 2.8 Å resolution. The new structural model supports and complements the original work and together, the two structures provide a framework for interpreting the attP mutagenesis results. Three key regions of Int-attP contacts appear to be the primary source of site specificity: the zinc ribbon domain, the RD-ZD linker, and the αE helix. Whereas the ZD and linker region requirements are well-supported by the structural models, the basis for αE preferences is less clear and may involve intermediates of the reaction with different protein and/or DNA conformations than that found in half-site structures. Together, these findings enhance our understanding of serine integrase recombination and provide important data for future efforts to engineer serine integrase systems.
Results
Energetics of Int binding to attP
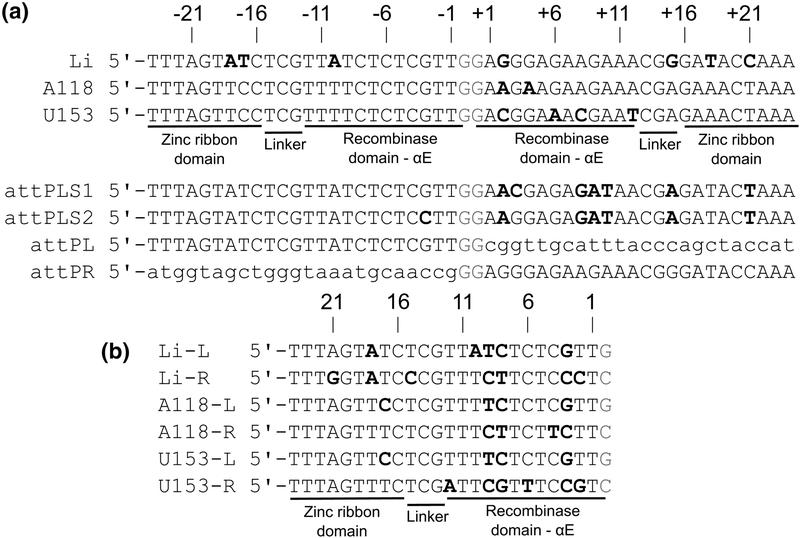
The attP sequence from the Listeria innocua prophage is shown in Fig. 2, with the closely related Listeria phage A118 and U153 attP sites for comparison. We determined the affinity of LI Int (Int) for its attP site using an electrophoretic mobility shift assay (EMSA) with endlabeled 116-bp DNA fragments (Fig. 3A). The data could be fit by a simple isotherm, with Kd=6.9±0.7 nM for the binding conditions described in Materials and Methods. Like the A118, Bxb1, and φC31 integrases, LI Int binds to attP as a dimer, with no evidence for step-wise binding of Int subunits [28,30,31]. We also examined binding of Int K362A, a coiled-coil (CC) mutant that is defective at intermolecular attP x attB recombination and carries out low levels of intramolecular attL x attR recombination in the absence of the phage encoded RDF [26]. The CC substitution results in higher affinity binding to attP, with Kd=1.1±0.1 nM (Fig. 3B). The implications of this observation are discussed below.
Figure 2.
Listeria attP sequences. A) Alignment of attP sequences from Listeria innocua prophage (LI), bacteriophage A118, and bacteriophage U153. The numbering scheme shown is used throughout the text. Approximate integrase domain interaction boundaries are indicated. Bold font indicates differences from the consensus sequence. attPLS1 is a symmetrized version of LI attP where the left half-site is duplicated. attPLS2 differs at position 3, where the nucleotide found in the right half-site of attP is symmetrized. Changes from wildtype attP in attPLS1 and attPLS2 have bold font. attPL and attPR contain the left and right half-sites of LI attP, respectively, with the other half-site replaced by an unrelated sequence. B) Alignment of the Listeria attP half-sites shown in panel A. Bold font indicates differences from the consensus sequence. In both A and B, the central crossover dinucleotide is colored red.
Figure 3.
Binding of LI Int to its attP site. A) Int binding to the attP site. B) Int K362A binding to attP. Lys362 is in the coiled-coil motif of Int and the alanine substitution weakens coiled-coil interactions. C) Int binding to attPL, the left half-site of attP. D) Int binding to attPR, the right half-site of attP. Example electrophoretic mobility shift assays (EMSAs) with 32P-labeled DNA probes and simple binding isotherm fits to the data are shown for each. We did not observe formation of higher order complexes in these experiments. Error bars are standard deviations from three independent titrations. Global fits to Kd were carried out with all measured data.
We next measured Int binding to probes where one of the two half-sites was replaced with an unrelated sequence. The attPL probe contains an intact left half-site and the attPR probe contains an intact right half-site (Fig. 2A). Int binds with higher affinity to attPL than attPR, and both are weaker than Int binding to attP (Fig. 3C,D). The similar electrophoretic mobilities of the Int•attPL and Int•attPR complexes compared to Int•attP suggest that Int dimers are bound in both cases, as demonstrated for Bxb1 Int binding to similar half-site probes [30]. One of the two subunits in the Int dimer binds to the attP half-site in these experiments, where the other is presumably bound non-specifically to the flanking DNA.
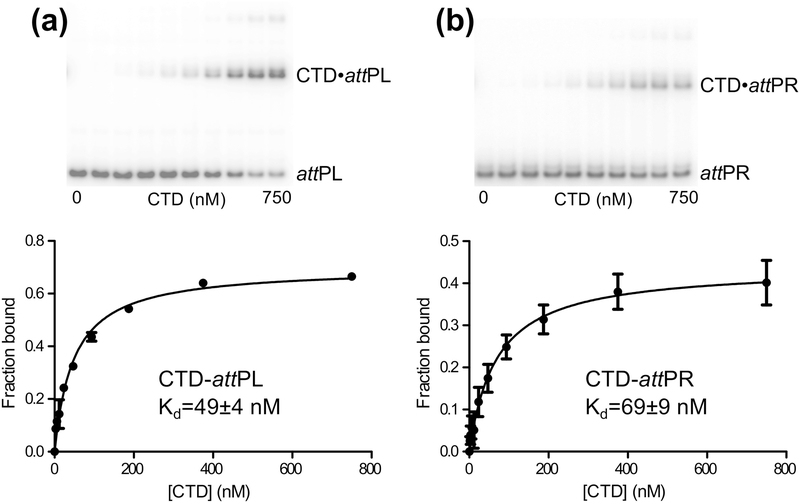
Finally, we measured binding of the Int CTD to attPL and attPR (Fig. 4). The CTD construct includes the same residues (133–452) used in the CTD•attPL crystal structure, but lacks the C-terminal hexahistidine tag that was used in that work [25]. The CTD binds to the attP half-sites with affinities weaker than full-length Int, but stronger than those previously reported for the A118, Bxb1, and φC31 integrases [23,28,32]. However, those studies were performed prior to knowing the structure of the CTD, and the protein constructs used were missing parts of the N-terminal end of the CTD that make numerous DNA contacts in the LI Int•attPL crystal structure. As expected, the LI Int CTD binds with higher affinity to attPL than to attPR, although the difference in binding energies is less than observed for full-length Int binding to the same sites. Some higher order complex formation was observed at higher CTD concentrations. These species may be CTD dimers binding to the half-site DNA, since we have previously shown that this domain dimerizes with a Kd of 20 μM and small amounts of dimer would be expected at the higher concentrations in the binding titrations [26]. A summary of LI Int-DNA binding affinities and standard free energies is given in Table 1.
Figure 4.
Binding of LI Int CTD to the attP half-sites. A) CTD binding to the left half-site, attPL. B) CTD binding to the right half-site, attPR. Example EMSAs with 32P-labeled DNA probes and simple binding isotherm fits to the data are shown for each. Error bars are standard deviations from three independent titrations and global fits were carried out in each case with all measured data.
Table 1.
LI Int-attP binding affinities
| Integrase | att site | EMSA Kd (nM) | ΔG° (kcal/mol), 30°C |
|---|---|---|---|
| Int | attP | 6.9 ± 0.7 | −11.3 |
| Int K362A | attP | 1.1 ± 0.1 | −12.4 |
| Int | attPL | 17.1 ± 1.0 | −10.8 |
| Int | attPR | 64.4 ± 7.2 | −10.0 |
| CTD (133-452) | attPL | 48.6 ± 3.8 | −10.2 |
| CTD (133-452) | attPR | 68.7 ± 9.4 | −9.9 |
New structure of a LI Int CTD•attPL complex
The previously determined crystal structure of LI Int CTD•attPL contained four independent copies of the protein-DNA complex in a cubic lattice, where non-crystallographic symmetry (NCS) averaging led to high quality electron density maps despite the modest resolution of 3.2 Å [25]. In hindsight, one of the structural features of that complex that made crystallization challenging was the presence of a flexible coiled coil (CC) motif that extends from the zinc ribbon domain and must be accommodated by the crystal lattice. Since the CC motif is not involved in DNA binding, we reasoned that a ΔCC CTD mutant might crystallize more readily when bound to attPL and the crystals might diffract to higher resolution.
We were able to grow orthorhombic crystals of LI Int CTDΔCC•attPL with one complex in the asymmetric unit that diffract to 2.8 Å, using a ΔCC construct that we previously described [25]. The structure was determined by molecular replacement and refinement rapidly converged to give the statistics summarized in Table 2. The CTDΔCC-DNA interface is similar in nearly all respects when compared to the CTD•attPL complex but provides an independent, higher resolution model (R.M.S.D of 1.05 Å for Cα carbons). The primary difference between the structures involves the αE helix, which binds to the minor groove near the center of attP (indicated in Fig. 1). In the CTDΔCC•attPL complex, the first four residues of this helix (Arg133-Met136) are poorly ordered and we do not observe the intercalation of Met136 into the base step at attP positions −1 and −2 that was seen with the CTD•attPL structure. Since the structures are so similar, we will not describe the CTDΔCC-DNA interface in detail, but instead point out where the new structure aids in our interpretation of attP mutant activities. Closeups of the CTDΔCC•attPL interface are shown in Fig. 5, with the docking of an α-helix from the recombinase domain into the major groove illustrated with electron density in Fig. 5A. This region is discussed in more detail below. In the results and discussion that follow, our structural interpretations refer to both the CTDΔCC•attPL and the CTD•attPL structures, unless otherwise indicated.
Table 2.
Summary of X-ray data processing and refinement statistics. Statistics for the highest-resolution shell are shown in parentheses. Rmerge = Σhkl Σi | Ii(hkl) - <I(hkl)>| / ΣhklΣi Ii(hkl).
| PDB code | 6DNW |
| Beamline | APS 24-ID |
| Wavelength (Å) | 0.97919 |
| Temperature (K) | 100 |
| Resolution (Å) | 40–2.85 (2.95–2.85) |
| Space Group | P212121 |
| Unit cell (Å) | a=48.3, b=57.1 c=167.9 |
| Total reflections | 34,318 (4,973) |
| Unique reflections | 11,185 (1,082) |
| Multiplicity | 3.0 (3.1) |
| Completeness (%) | 97.5 (98.7) |
| Mean I/sigma(I) | 14.9 (2.3) |
| Wilson B-factor (Å2) | 53.2 |
| R-merge (%) | 5.7 (41.4) |
| Reflections used for R-free | 578 (5.2%) |
| R-work | 0.219 (0.379) |
| R-free | 0.257 (0.380) |
| Number of non-hydrogen atoms | 3,018 |
| protein & DNA | 3,008 |
| zinc ion | 1 |
| water | 9 |
| Protein residues | 231 |
| RMS(bonds) | 0.003 |
| RMS(angles) | 0.54 |
| Ramachandran favored (%) | 96.0 |
| Ramachandran allowed (%) | 4.0 |
| Ramachandran outliers (%) | 0.0 |
| Rotamer outliers (%) | 0.0 |
| Average B-factor (Å2) | 56.7 |
| Protein & DNA (Å2) | 56.7 |
| Solvent (Å2) | 34.3 |
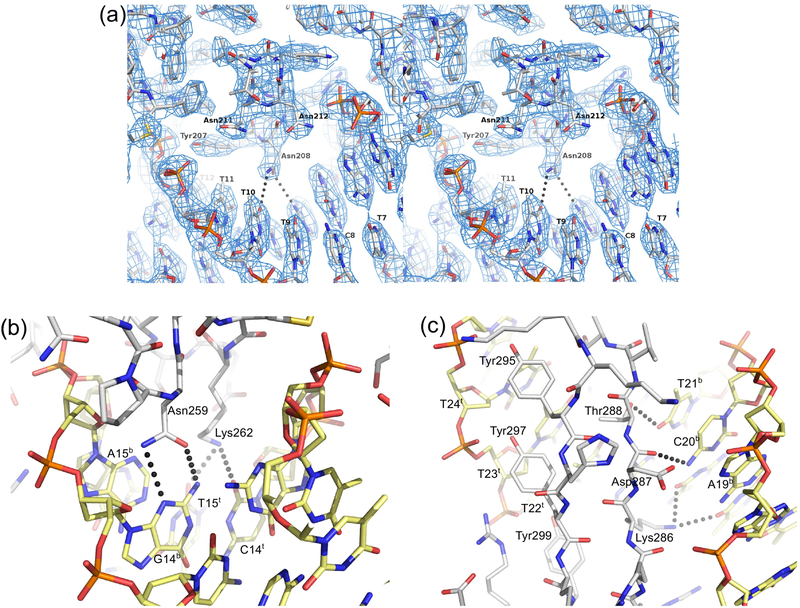
Figure 5.
LI Int CTDΔCC•attPL structure. A) Stereo view of recombinase domain-major groove interactions. An α-helix from the recombinase domain is shown docked into the attP site major groove. DNA residue labels refer to the A118 attP left half-site shown in Fig. 2B. The Asn208-DNA long-range hydrogen bonds (3.5–3.7 Å) shown are the only polar interactions made in this major groove interface. No water-mediated interactions are observed in this region. The only other direct contact to bases involves Tyr207, which makes van der Waals contact with the C5-methyl group of T11. Electron density from a weighted 2Fo-Fc map is shown contoured at 1.4 σ. B) Minor groove interactions between the Int linker region and attP residues 13–15. The specific interaction between Asn259 and G14b and the pyrimidine-specific interactions involving Lys262 are shown. C) Major groove interactions between the zinc ribbon domain β-hairpin and attP residues 19–24. Three tyrosine residues make extensive non-polar contacts to the C5-methyl groups of residues T22t-T24t and to the DNA backbone. Polar interactions involving Lys286, Asp287, and Thr288 are shown. Pymol was used to make this figure [48].
A symmetric attP substitution library
To analyze the effects of attP substitutions, we constructed a plasmid library of mutant attP sites where all possible symmetric substitutions were introduced in positions 1–24 of the halfsites (see Fig. 2 for attP numbering). This covers the Int-DNA contact region observed in the LI CTD•attPL and CTDΔCC•attPL crystal structures and is consistent with the requirements for efficient A118 Int recombination [28]. Since our strategy involved symmetrizing individual positions, we asked whether an attP variant with half-sites arranged as perfect inverted repeats on either side of the central GG dinucleotide was functional. This is a non-trivial question, since Mandali et al. [28] have shown that a symmetric attB generated by duplication of the left half-site is poorly active in attPxattB (PxB) recombination. We therefore tested a symmetric attP site containing duplications of the left half-site (attPLS1) for intermolecular PxB integration (Fig. 6). In this assay, a 120-bp attB is integrated into a linearized 3 kb plasmid containing attPLS1, generating 1.4 and 1.7 kb products (Fig. 6A). Despite the binding preference for attPL over attPR by both full-length Int and Int CTD, duplication of this half-site results in a weaker substrate for the integration reaction compared to attP (Fig. 6BC). Thus, PxB recombination does not strictly require an asymmetric attP site but sequence elements found in the weaker binding right half-site must make important contributions to integration activity.
Figure 6.
In vitro integration activities of symmetric attP variants. A) Schematic of the in vitro integration reaction. A short attB fragment is integrated into a linearized attP plasmid to produce attL and attR products. Arrows over the attachment sites indicate the relative orientation of the sites (as drawn in Fig. 2). B) Examples of time courses of reactions containing 6.5 nM attP, 150 nM attB, and 200 nM Int and incubated at 32°C. Reaction aliquots were quenched at the given time points and analyzed on agarose gels as described in Materials and Methods. C) Comparison of integration kinetics for wild-type and symmetric attP sites. Error bars are standard deviations based on three independent experiments.Errors smaller than the plotting symbol are not shown.
For each mutant substrate, the wild-type attP site has been modified so that the same nucleotide in both half-sites has been changed in the same way with respect to the 5’−3’ sequence, resulting in a symmetric substitution. A substitution in the left half-site top strand is paired with the complementary substitution in the right half-site top strand of attP (see Fig. 2A), since the half-sites are arranged as inverted repeats. We refer to individual attP mutants by the left half-site sequence at a given position (i.e., the first sequence shown in Fig. 2B), since this is the sequence for which we have supporting structural models. The mutant 2G, for example, introduces G in the left half-site at position −2 and C in the right half-site at position +2 of attP. The 2T substitution replaces only position +2 with A, since −2 is already T in wild-type attP. Individual residues referring to the top strand as shown in Fig. 2A are given with a ‘t’ superscript (e.g., −3Gt) and those referring to the complementary bottom strand are indicated with a ‘b’ superscript (e.g., −2Ab). We constructed the attP library using an R6kγ plasmid to facilitate testing the sites for their ability to undergo integration into a single attB site in E. coli, as described below. In all, 79 different attP mutants were generated (listed in Table S1).
In vitro integration of attP mutants.
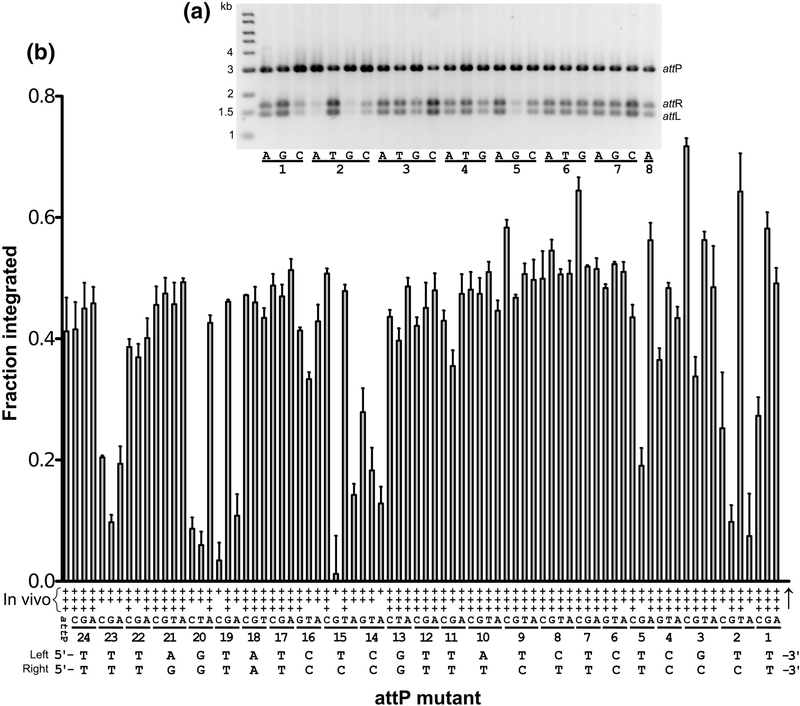
To test the ability of attP mutants to undergo intermolecular PxB recombination, we linearized the attP plasmids and measured the fraction of recombinant products generated when incubated with a small linear attB site and Int for 60 min at 32°C (Fig. 7). For wild-type attP, approximately half of the substrate is converted to product in 60 min with our reaction conditions and substrate concentrations (Fig. 6). Although reactions with supercoiled plasmid substrates are more efficient, we opted for the linear variant of this reaction in order to avoid complications resulting from differences in the amount of supercoiled vs. open circle forms and the presence of plasmid dimers in the library. Examples of reactions from a subset of the mutants are shown in Fig. 7A and the results from all attP mutants are summarized in Fig. 7B and Table S1.
Figure 7.
Integration activity of attP mutants. A) Results of in vitro integration for the first 24 of 79 mutants. The symmetric nucleotide substitutions associated with each mutant are indicated for the given position as defined in Fig. 2 and explained in the text. For example, the first three mutants tested are 1A, 1G, and 1C. Reactions were performed as in Fig. 6 but were quenched after 60 min. B) Summary of integration activities for all attP mutants. In vitro integration activity is given as the fraction of products formed in 60 min at 32°C. Error bars are standard deviations based on three independent experiments. In vivo integration activity was measured as the transformation efficiency of a replication-incompetent attP plasmid into an E. coli strain containing a single copy attB site as described in Materials and Methods. In vivo integration activities are indicated as + (<10% of wild-type attP), ++ (1060%), or +++ (>60%). The two wild-type attP half-site sequences are shown at the bottom, labeled ‘left’ and ‘right’. The crossover site is marked by a vertical arrow.
Of the 24 half-site positions, only three show a strong preference for a single residue. At position 2, T is preferred. In both CTD-DNA crystal structures, Lys140 from the E-helix is positioned in the minor groove where it could interact with either the −2 or −3 residues. In the CTD•attPL model, the interaction is with the −2Ab base, but in the CTDΔCC•attPL model, the interaction is with the −3Cb base. These interactions do not provide a satisfactory explanation for the 2T preference, since Lys140 appears well-positioned to interact with alternative residues at both the −2 and −3 positions. The −2 base-pair is also adjacent to a distortion in the DNA, where Met136 from αE intercalates between −0Gt and −1Tt in the CTD•attPL structure. It is not yet known if this interaction is functionally important for recombination activity; as noted above, we do not observe the intercalated methionine in the CTDΔCC•attPL structure. The second specific attP position is 14, where C is preferred. The linker connecting the recombinase and zinc ribbon domains of Int interacts in the minor groove at this location, where Asn259 makes an unusual double hydrogen-bonding interaction with −14Gb (Fig. 5B). The third specific site is at position 23, where T is preferred. The β-hairpin of the zinc ribbon domain lies in the major groove in this region, where Tyr295 makes van der Waals contact with the −23Tt methyl group (Fig. 5C).
Three half-site positions have a strong preference for either of two nucleotides. At position 15, T and C are preferred. Interactions between Lys262 from the inter-domain linker and O2 of either T or C in the minor groove is the most likely explanation, since this residue interacts with O2 of −15Tt in both structures (Fig. 5B). At position 19, G and T are preferred. Lys286 hydrogen bonds to O4 of −19Tt in the structural models; a similar interaction with O6 of −19Gt could be made. The same lysine side chain also interacts with N7 of -G20t (Fig. 5C). The third dual preference is at position 20, where A and G are most active. We noted a hydrogenbonding interaction between the Asp287 backbone carbonyl in the β-hairpin of the zinc ribbon domain and N4 of −20Cb in the CTD•attPL structure and assumed that it would be highly specific. The same interaction is present in the CTDΔCC•attPL structure described here (Fig. 5C). Our finding that 20A is also a good substrate is unexpected, since the −20Tb C5-methyl group is predicted to lie about 2.5 Å from the Asp287 backbone carbonyl. Indeed, we previously showed that the 20T mutant is defective for both integration and attP binding, suggesting that −20Ab disrupts the β-hairpin interaction [25]. It is not clear why the 20A mutant is active both in vitro and in vivo; we assume that a compensatory adjustment in the βhairpin backbone must be formed to avoid steric clash with the −20Tb methyl group.
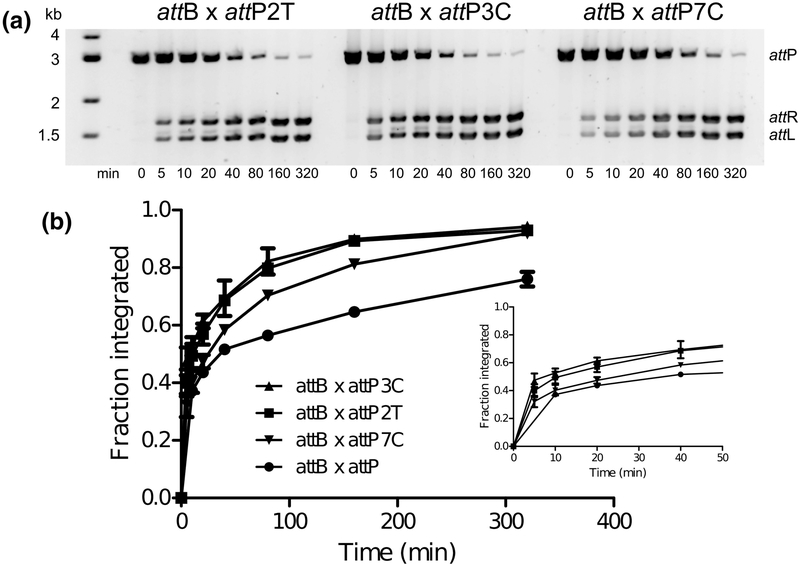
Two overall features from the results shown in Fig. 7 stand out. First, many attP positions are relatively insensitive to single, symmetric substitutions. Thus, despite the extended 50 bp contact region between Int and DNA in the Int•attP complex, specificity appears to be derived from just a fraction of the sequence. The second feature is that several symmetric substitutions lead to higher activity than the wild-type attP site. The 2T, 3C, and 7C mutants had the highest in vitro integration activity and were further investigated as described below.
In vivo integration of attP mutants
To determine if attP mutants have similar relative activities in vivo, we used an assay that we previously reported [25,26], where integration activity is scored as the transformation efficiency of an attP-containing R6kγ plasmid into a F’-attB strain expressing Int. To provide an internal control, we co-transformed a compatible 3 kB plasmid with a streptomycin resistance marker (see Materials and Methods). The results are summarized in Fig. 7b and Table S1. Although we have previously determined quantitative activities with this assay [26], we report qualitative results here due to the variation in plasmid topologies across the mutant library that we noted above. Mutant plasmids were scored as either weak (< 10% of wildtype), medium (10–60%) or strong (> 60% wild-type). Overall, there is good agreement between the in vivo and in vitro integration activities. The only discrepancy is the 2G mutant, which is more active in vivo than we would have expected from its in vitro activity.
The properties of two attP mutants differ from what we originally reported. We found here that the 21T and 13T substitutions are competent for Int binding, in vitro integration, and integration in E. coli. Rutherford et al. [25] reported weak in vivo activity for both mutants, which we believe was the result of not passaging a subset of the mutant plasmids tested (13T, 14A, 15G, and 21T) through a methylation-competent strain that prevented restriction by the tester strain CSH142 (which is not hsdR). This issue was avoided in subsequent work by performing integration assays in strain BW25113 (which is hsdR).
Int binding to attP mutants
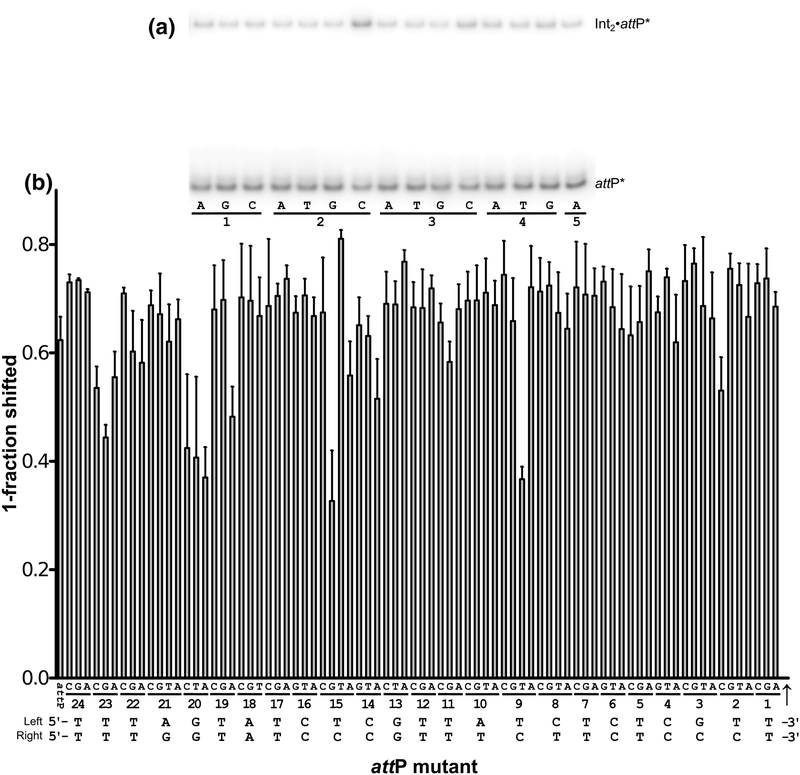
The simplest interpretation of the integration activities shown in Fig. 7 is that changes to the attP sequence either reduce, enhance, or do not affect binding of Int and the observed integration activities reflect those differences in binding affinity. However, we and others have found that some att site mutations can have dramatic effects on recombination but little effect on att site binding [23–25]. To determine if this is the case for any of the LI attP variants, we tested the ability of Int to bind to each attP mutant. We carried out these experiments in a competition format, where an attP mutant DNA fragment was tested for its ability to compete with a radiolabeled attP probe for Int binding. For example, the first six attP mutants shown in Fig. 8A bind efficiently to Int, resulting in depletion of the shifted label. The 2C mutant is bound with less affinity by Int, resulting in less depletion of the shifted label. The competition experiments for a subset of the library are shown in Fig. 8A, with results for all mutants summarized in Fig. 8B and Table S1.
Figure 8.
Binding of Int to attP mutants. Relative binding affinities were measured by the ability of an attP mutant PCR fragment (32 nM) to compete with a 32P-labled attP fragment (<1 nM) when incubated with 16 nM Int. Mutants that are weakly bound by Int will result in increased shifted label in an EMSA. Mutants that are tightly bound by Int will result in a decrease in shifted label. A) EMSA results for the first 15 attP mutants, where 2C can be readily identified as bound with less affinity by Int. B) Summary of competition binding by all attP mutants. Error bars are standard deviations resulting from 4–5 independent experiments for each mutant. The two wild-type attP half-site sequences are shown at the bottom, labeled ‘left’ and ‘right’. The crossover site is marked by a vertical arrow.
Of the nine attP mutants most defective for Int binding (2C, 9T, 14A, 15G, 19A, 20A, 20T, 20C, and 23G), all but two (9T and 20A) also showed defects in integration activity. As discussed above, the 20A substitution results in an active attP site, contrary to our expectations. This mutant is defective in Int binding, however, which is consistent with a structure-based prediction of the effects of this substitution. The 9T mutant symmetrizes the T found in the left half-site at −9 and like the 20A mutant, retains strong integration activity despite the lowered Int binding affinity. Interestingly, all three Listeria attP sites have T at −9 and G at +9 (Fig 2) and the symmetric 9C mutant is highly active in both integration and Int binding. The base-pairs at −9 and −10 interact with the Asn208 side chain in a network of longrange hydrogen bonds (3.5–3.7 Å). This interaction, shown in Fig. 5A for the CTDΔCC•attPL structure, is the only example of direct polar contacts between the recombinase domain (RD) and bases in the major groove of attP. As discussed further below, the RD places an α-helix in the major groove of the attP site, but provides only limited direct recognition of the DNA sequence.
In general, most residues that appear defective in attP-binding also show integration defects. The converse, however, is not necessarily true. Several attP mutants that are defective in integration are not severely affected in their ability to bind Int. Examples include 1C, 2A, 2G, 5G, 14T, 14G, and 19C. We did not further investigate these mutants to determine which step of the integration pathway is being disrupted.
attP sites with enhanced integration activity
Several attP mutants showed higher integration activity compared to the wild-type site. To further explore these observations, we measured integration time courses for the three most active mutants: 2T, 3C, and 7C (Fig. 9). Each of these sites leads to rapid product formation, with detectable attL and attR within seconds of adding Int to the reactions (Fig. 9A). The 2T and 3C mutants are similar to one another in their reaction kinetics, whereas 7C starts somewhat slower. The 2T mutant symmetrizes the nucleotide found in the left half-site of attP, whereas the 3C mutant symmetrizes the nucleotide found in the right half-site (Fig. 2). These observations provide a plausible explanation for why the tighter binding left half-site leads to a less functional attP when symmetrized: the favored +3G residue in the right halfsite is lost. To test this idea, we made the 3C substitution in the context of the attPLS1 sequence to generate attPLS2 (Fig. 2A). As shown in Fig. 6, this change leads to a symmetric attP that is highly active at integration, being converted to 50% products in the first five minutes of incubation. Asymmetry in the attP sequence is therefore not required for efficient integration. Nonetheless, the −3G/+3G asymmetry has been maintained in each of the LI, A118, and U153 attP sequences (Fig. 2A).
Figure 9.
LI attP substitutions resulting in high integration efficiencies. In vitro integration time courses were carried out as in Fig. 6. A) Examples of reaction time courses for the 2T, 3C, and 7C mutants. B) Integration kinetics of hyperactive mutants vs. wild-type attP. Error bars are standard deviations based on three independent experiments. The inset graph shows early time points. Rapid product formation is evident from the zero time point, where reactions were quenched within 1–3 sec after initiation.
The structural basis for a C preference at position −3 is not obvious from our existing structural models, where the less favored G is present. As noted above, the two structures differ in the positioning of Lys140 in this region; in the higher resolution CTDΔCC•attPL model, Lys140 interacts with O2 of the −3Cb base, whereas the same residue interacts with N7 of the 2Ab base in the CTD•attPL model. In both cases, the side chain is well ordered but adopts a compact, poorly extended conformation. It is possible that −3C would allow a more productive interaction involving an extended Lys140, where bridging interactions could occur between exocyclic oxygen atoms in both the −2T and −3C base-pairs. Indirect readout of this region of the attP site could also explain the preferences observed, where sequence-dependent DNA deformations may be required to facilitate cleavage and strand exchange. Regardless of the detailed nature of the interactions and helical conformations involving −3C vs −3G, it seems likely that the functional importance of this position involves a later stage of the reaction, since the 3G and 3C mutants are both bound by Int with high affinity. The residue corresponding to Lys140 in the φC31 integrase is also lysine and C is found at positions −3t and +3b in both the attP and attB sites, suggesting that similar Int-DNA interactions may be used in that system.
The highly active 7C substitution is surprising, given that the wild-type attP site has T in all six Listeria attP half-sites at this position (Fig. 2B). The −7 base-pair does not interact with Int in the structural models, nor are there obvious water-mediated interactions that could be made.
Flexibility in attP sequences
The in vitro integration activities for all attP mutants are illustrated schematically in Fig. 10A, with the nucleotide representation of six Listeria attP half-sites shown in Fig. 10B for comparison. A striking feature of these results is the large number of sites that are tolerant to nucleotide substitutions. In many cases, these positions show no variation among the Listeria attP half-sites that have been characterized thus far. There are only three regions in the attP site that show strong sequence preferences. The first is positions 19–23, where the zinc ribbon domain binds (Fig. 5C). The second is positions 14–15, which is where the short linker peptide connecting the recombinase and zinc ribbon domains interacts (Fig. 5B). In both cases, there is good agreement between contacts observed in the CTD•attPL and CTDΔCC•attPL half-site crystal structures and the specificity inferred from integration and DNA-binding efficiencies. The third region is at positions 2–3, where the αE helix from Int interacts in the minor groove of the site. Here, a structural explanation in the context of attP site binding is less clear.
Figure 10.
Schematic representation of LI attP sequence preferences. A) In vitro integration activity as a function of sequence position, where letter size is proportional to activity. B) Half-site sequence frequency based on the alignment shown in Fig. 2B for six Listeria attP half-sites. Plots were made using WebLogo [49].
Discussion
In this work, we have investigated the binding energetics and sequence requirements of the LI attP site. The Int CTD binds about 10-fold weaker to the attP half-sites compared to Int binding to the full site, which is not the the result one would expect if the two half-site binding energies were additive. In that case, an affinity of ~4 pM (−20 kcal/mol) would be expected for Int-attP binding, based on the measured CTD binding affinities shown in Table 1. The inability of a simple half-site avidity model to explain attP binding is also evident from the full-length Int binding data. Based on the binding energies of Int to attP, attPL, and attPR, we would expect energies of 6.0, 5.2, and 4.8 kcal/mol for binding to the left, right, and non-specific half-sites of our DNA probes, respectively, if half-site contributions were additive. These estimates imply affinities of 47 and 177 μM for the left and right half-sites, in strong disagreement with the CTD binding data. Similar observations were reported for the mycobacteriophage Bxb1 integrase, leading to the suggestion that some of the half-site binding energy is offset by reorganization of the Int dimer as a prerequisite to binding [30].
Since the actual LI Int-attP binding energy is −11 kcal/mol, +9 kcal/mol must be expended to allow an Int dimer to bind attP. LI Int forms stable dimers in the absence of DNA, with a monomer-dimer Kd ≤ 32 nM [26]. We previously suggested that interactions between coiledcoil domains contribute to this dimer interface, since deletion of the CC motif results in Int dimers that are over 100-fold weaker, with Kd = 5.7 μM [26]. Thus, disruption of CC-mediated interactions within an Int dimer may be the primary remodeling event required for the two CTDs to bind the attP half sites [33]. Our estimate of 20 μM for the CC dimerization Kd [26] corresponds to a standard free energy of −6.5 kcal/mol at 30°C, which would account for a large fraction of the 9 kcal/mol discrepancy. Distortions of the DNA and the αE helix near the center of attP may also require free energy to be expended upon formation of the Int2•attP complex.
If the above model is correct, then Int mutants that weaken the interaction between CC motifs in an Int dimer should result in tighter binding to attP, since disruption of CTD interactions will be easier and the CC motifs cannot interact once Int is bound to attP (see Fig. 1). This is indeed the case for the Int K362A mutant, which forms weaker CTD dimers than wild-type CTD [26] and binds 6–7 fold tighter to attP compared to wild-type Int (Table 1). The Y369A substitution also weakens the ability of CC motifs to interact and reduces in vitro integration activity [26]. We found that this Int variant binds with similar affinity to attP as does K362A (data not shown).
Our analysis of LI attP mutants revealed three regions that are most sensitive to substitutions. These regions correspond to the binding sites for the ZD, the RD-ZD linker, and the αE helix. A rationale for the sensitivity of the ZD-binding region (positions 16–24) is clear from the two independent structural models of the CTD•attPL complex because of the direct Int-DNA interactions that can be observed. The ZD-binding region in LI attP corresponds to the flanking sequences in the Bxb1 attP site that are the most sensitive to mutation in the entire site [23]. Substitutions at Bxb1 positions 18, 19, 20, and 22 (LI numbering) are mildly defective for Int binding but severely defective in recombination. For LI attP, positions 19, 20, and 23 are most sensitive in the ZD-binding region, suggesting that LI and Bxb1 integrases use a similar mode of DNA recognition involving the β-hairpin of the ZD.
Other investigators have also observed that mutants in the outermost regions of attP and attB sites can be bound by Int with reasonable affinity, yet be defective for recombination [22–24]. The favored hypothesis to explain these results is that a subtle conformational change somehow makes the Int-bound sites less suitable for partner association and recombination. In the context of current structural and mechanistic models, mis-positioning of the CC motifs could be the functional consequence of these changes, but it is not yet clear why such effects would in some cases be manifested in later stages of the reaction rather than simply the association of sites [23].
Interactions involving the RD-ZD linker (positions 13–15) also have straightforward structural interpretations for LI attP. Positions 14 and 15 are most sensitive, but changes to the corresponding region for Bxb1 attP were not found to affect binding or recombination, at least for the single substitutions tested [23]. The linker-binding region of the attP site may therefore only be used by some serine integrases for specific site recognition and discrimination. This is understandable, given that the LI Int interactions in this region involve a highly specific asparagine-guanine pairing that is not typical of generally less specific minor groove contacts. An additional role for this region of the Listeria phages could be to ensure that attP is not mistaken for attB. Since the favored model for Int-attB binding is incompatible with the linker-DNA interaction shown in Fig. 5B, this region may contribute to disfavoring an attB binding configuration on an attP sequence.
We were particularly interested in identifying sequence preferences located in the αEbinding minor groove of the attP site (positions 0–3). The Bxb1 attP site is sensitive to substitution at positions ±3 (LI numbering), with Int-binding and recombination both severely affected [23]. In the φC31 attB site, positions ±2 are sensitive to substitution [24], which is the same result we found for LI attP. These sequence requirements may be due in part to direct αE-DNA interactions, some of which could occur in the initially formed Int-DNA complexes. Other αE-minor groove interactions may occur later in the reaction pathway and could be associated with DNA distortions required for strand cleavage. Indirect readout of the αE binding region could also play a role throughout the recombination pathway, where sequencedirected shape and deformability may be important.
In contrast to the zinc ribbon domain, the recombinase domain (binding at positions 3–12) appears to contribute very little to specific recognition of the LI attP sequence. Like the small helix-turn-helix domains found in the small serine recombinase enzymes [2], the RD places an α-helix in the major groove of the DNA substrate (shown in Fig. 5A). For LI Int, however, there are only two direct contacts involving this helix and the interactions that are present appear to accommodate nearly any single substitution that is made. In addition to the Asn208 interactions discussed earlier, Tyr207 forms van der Waals contact with the C5-methyl group of -T12t (also visible in Fig. 5A). Singh et al. found little evidence for sequence discrimination in the corresponding region of the Bxb1 attP sequence [23] and in a study of φC31 attB mutants, Gupta et al. found that this region of the site was relatively tolerant to substitutions [24].
Xiaolai et al reported similar results for the phage φBT1 attB site [22]. Although there are currently no structures known for a serine integrase bound to an attB sequence, current models suggest that the protein-DNA interactions in the αE-RD region of attB should be similar to attP [15,25,33]. Thus, there is now considerable evidence that the RDs of serine integrases do not, in general, provide a high level of specific recognition of attachment sites. This suggests that an opportunity may exist for introducing new serine integrase specificity through engineering of the RD helix-turn-helix motif.
Overall, 14 of the 24 positions in the attP half site can be substituted symmetrically with any alternative while retaining at least 75% of wild-type attP recombination activity (positions 4, 6–8, 10–13, 16–18, 21–22, and 24), leaving ten positions with sequence preferences (Fig. 10). Several of the ten specific positions have more than one preferred sequence. There is therefore considerable flexibility present in the attP sequence and many alternative sites are likely to be functional for integration with no changes required of the integrase protein. Our findings are consistent with the identification of multiple φC31 attP-like sequences in the human and mouse genomes identified by Calos and colleagues [21,34]. In those studies, φC31 Int was shown to integrate attB-containing plasmids into a number of ‘pseudo-attP’ sites, which resembled the wild-type site in the αE and RD binding positions (2–12), but showed little similarity in the flanking sequences. Thus, many of the key residues conferring specificity in the ZD-binding region of φC31 attP can be absent and the site still remains functional at some level. A caveat of this interpretation is that we are assuming that the LI Int structural models can adequately predict the nature and position of ZD-DNA interactions in the φC31 system.
One of the questions raised in this work was whether asymmetry plays a functional role in the attP site. Early studies of serine integrases showed that the central dinucleotide alone determines the attP-attB site alignment that will lead to integration, indicating that the half sites are functionally equivalent for the PxB reaction [35,36]. However, duplication of a halfsite to generate a symmetric full site does not always result in a functional substrate, as Mandali et al. showed for A118 attB [28]. We found that a symmetric LI attP generated using the tighter binding left half-site (attPLS1) was less active than attP, but still functional. A symmetric 3C substitution in attPLS1 resulted in a symmetric site (attPLS2) that is a much better substrate than attP. Thus, fully symmetric attP sites can be constructed that integrate with high proficiency.
Despite the lack of a requirement for attP asymmetry, both Bxb1 Int and the Listeria Ints have different DNA-binding affinities between left and right attP half-sites [23,28]. The differences between half-sites could simply be a consequence of regulatory sequence constraints imposed by the phages. LI Int binds more tightly to attPL than attPR and we can rationalize these observations from the mutational studies reported here. Based on binding affinity, the primary difference between the LI attPL and attPR half-sites is at position 2, where −2T is favored and +2G is defective. Based on integration efficiency, LI attPL has two favored (−2T and −9T) and one unfavored (−3G) sites and attPR has two unfavored (+2G and +9G) and one favored (+3G) sites. In forming the attPLS1 symmetric site, we duplicated the unfavorable −3G position, which has a strongly negative impact on recombination. Thus, asymmetry in LI attP involves both favorable and unfavorable substitutions on both half-sites. A118 Int also binds better to the left vs the right A118 attP half-site, but our results do not provide a clear explanation. The A118 attP site is symmetric at position 2, with the favored 2T sequence. It is possible that our competition DNA-binding assays are not sufficiently sensitive to identify subtle differences in affinity at positions 3, 4, 8, 9, and 17, where the A118 attP site contains asymmetric residues.
Many substitutions in the LI attP site result in a better substrate for the attP x attB reaction. These results suggest that there is an opportunity for considerable improvement in the efficiency of the integration reaction for LI Int and perhaps for other serine integrases, by engineering the attachment sites. It will be important and interesting to determine if higher efficiency attP and attB sequences are compromised in any way with respect to the stability of attL and attR and if the RDF-mediated excision reactions are equally efficient. Although serine integrase RDFs have been shown to bind the integrase protein and not the attachment site sequences [29,37–39], a role for weak RDF-DNA interactions that could potentially be affected by sequence changes has not yet been ruled out. Conformational changes resulting from an altered attP sequence could also affect RDF interactions indirectly.
For some of the attP substitutions we studied, structural models of the Int-DNA interface were valuable for interpreting the results. However, the CTD•attP and CTDΔCC•attP half-site crystal structures do not capture the geometry of the intact αE helices when an Int dimer is bound to the full attP site. Even if a pre-synaptic Int•attP structure were available, the model would still likely provide little information about how the Int-αE interface changes as the reaction progresses through synapsis, cleavage, and subunit rotation. The saturation mutagenesis data we describe here provides the overall attP sequence requirements for efficient integration, but further studies will be required to identify the defective steps in the reaction pathway for the integration defective, but DNA-binding competent substitutions. We found that it was important to evaluate all possible substitutions at each site, since some positions have dual preferences that could be missed with a single substitution strategy.
Protein engineering to modify sequence specificity has been a major field of research for a number of years, with important progress reported for zinc fingers, meganucleases, TAL effectors, and other DNA-modifying enzymes [40]. The LI attP sequence preferences reported here will be useful in similar efforts for the serine integrases. A complementary approach will be to take advantage of the modular nature of the serine integrases by exchanging DNA-binding units with those having alternative specificities. This has already been done successfully for the small serine recombinases, where the HTH DNA-binding domains have been substituted with zinc finger modules [41]. The serine integrases have two such modules that can be swapped: the recombinase domain and the zinc ribbon domain. Since the serine integrase coiled-coil motifs play a crucial role in facilitating integration and controlling directionality of recombination, alternative or engineered RD-ZD modules may be required in order to maintain the desirable properties of these enzymes. Understanding the sequence requirements of serine integrase attB sites is equally important in this endeavor and is the subject of ongoing research in our laboratory.
Materials and methods
Strains and reagents.
Strain BW25113 (F-, Δ(araD-araB)567, lacZ4787(del)::rrnB-3, LAM-, rph-1, Δ(rhaD-rhaB)568, hsdR514) was obtained from the Coli Genetic Stock Center (Yale University). Buffer pH values were measured at 25°C. Biochemical reagents were obtained from Sigma-Aldrich or Fisher Scientific. The protein sequence for the Listeria integrase used in this work is given in GenBank entry CAC97653.
Purification of LI Int and LI Int truncations.
Full-length Int and Int CTD (residues 133–452) were expressed and purified as described [26]. Int CTDΔCC (residues 133–452/Δ342–416) was expressed and purified as described [25]. Concentrations were determined by UV absorption, using calculated extinction coefficients of 65.4 mM−1cm−1 for Int, 50.0 mM−1cm−1 for Int CTD, and 43.8 mM−1cm−1 for Int CTDΔCC [42].
Electrophoretic mobility shift assays.
DNA probes were 116 bp EcoRI fragments containing attP or attP variant sequences that were end-labeled with α−32P-dATP and Klenow fragment DNA polymerase (NEB) and purified on Centri-Spin-20 columns (Princeton Separations). Binding reactions of 20 μL contained < 0.5 nM DNA probe and varying amounts of integrase in binding buffer (20 mM Tris hydrochloride, pH 7.4, 150 mM potassium chloride, 2 mM dithiothreitol (DTT), 25 μg/ml bovine serum albumin (BSA), 25 μg/ml salmon sperm DNA, 5% glycerol), and were incubated for 30 min at 32°. Reactions were separated by electrophoresis at 14 V/cm on 6% polyacrylamide (37.5:1) using Tris-glycine buffer, pH 8.3, at 15°C using a Hoefer SE600 system and a recirculating water bath. Integrase titrations were prepared by serial 2-fold dilution in binding buffer followed by mixing with an equal volume of DNA probe in binding buffer. Dried gels were scanned using a Typhoon imager (GE) and quantitated using Image Studio Lite (LI-COR). Binding data were fit to the simple binding isotherm f = fmax[Int]/([Int] + Kd), where f = fraction bound = [Int•DNA]/([Int•DNA] + [DNA]) using GraphPad Prism (GraphPad Software). All titrations were repeated three or more times and estimated errors in Kd values were based on global fitting to all experimental data.
Structure Determination.
The DNA construct used for co-crystallization with LI CTDΔCC is identical to that used for the CTD•attPL structure [25] and contains the left half-site of phage A118 attP. The sequence differs from LI attPL in three positions (10, 17, and 18), each of which was found to be insensitive to substitution (this work). Crystals of LI CTDΔCC•attPL were obtained by hanging drop vapor diffusion at 21oC with 30 μM complex in 100 mM Tris HCl, pH 7.5, 200 mM ammonium sulfate, and 30% (w:v) polyethylene glycol 3350. Crystals were cryo-protected in crystallization buffer plus 30% glycerol, flash-frozen, and stored in liquid nitrogen prior to diffraction experiments. Diffraction data were collected at the Advanced Photon Source NE-CAT beamline 24ID and data were processed using XDS [43]. The structure was determined by molecular replacement using PHASER [44], with a truncated model based on the LI Int CTD structure (PDB code 4KIS), revealing 1 complex in the asymmetric unit. The structure was refined initially using CNS [45] and fit using composite omit maps in COOT [46] and in the final iterations using PHENIX [47]. A summary of diffraction data and refinement results is given in Table 2. Coordinates and structure factors have been deposited in the Protein Data Bank with code 6DNW.
Construction of attP mutant plasmids.
The wild-type LI attP plasmid (pGV2345) contains the 56-bp attP sequence deduced from the Listeria innocua CLIP11262 genome (GenBank NC_003212), flanked by 21-bp and 18-bp primer sequences. The attP cassette was cloned into the BamHI and HindIII sites of pFBR6kamp, an R6kγ plasmid containing an ampicillin marker (G.V., unpublished). This plasmid can only replicate in strains containing the pir gene. Mutants were generated by inverse PCR mutagenesis using long, 5’-phosphorylated forward primers (IDT ultramers) containing the sequence changes and a short, unphosphorylated reverse primer. After ligation of the PCR product and treatment with DpnI (NEB), plasmids were transformed into strain EC100D pir-116 (Epicentre) and sequenced.
In vitro integration reactions.
Intermolecular attP x attB integration reactions were carried out between a linearized attP plasmid and a 120-bp attB site generated by PCR. The attP plasmids were linearized by BglII digestion, resulting in the 56 bp attP site flanked by 1569 bp on the left and 1350 bp on the right (using orientation defined in Fig. 2). attB was amplified from pGV2693, a pIDTSmartKan (IDT) attB plasmid constructed as described for pGV2345, using primers directly flanking the site. attB fragments were purified using silica spin columns and quantified by UV absorption following exhaustive column washing to remove traces of guanidine. Concentrations were verified by agarose gel electrophoresis and comparisons to mass markers. Integration reactions contained 6.5 nM attP substrate, 150 nM attB substrate, and 200 nM Int in integration buffer (25 mM Tris hydrochloride, pH 8, 100 mM potassium chloride, 50 mM sodium chloride, 1 mM spermidine hydrochloride, 5 mM magnesium chloride, 2.5 mM dithiothreitol, 5% glycerol), and were incubated at 32°C. For time courses, reaction volumes were 80 μL and aliquots of 10 μL were quenched with 2.5 μL 1% sodium dodecylsulfate, 0.02% bromophenol blue after various time intervals. For analyses of attP mutants, reaction volumes were 10 μL and were quenched after 60 min. Reaction products were separated on 1.2% agarose LE gels using 0.5X Tris-borate-ethylenediamine buffer, stained with GelStar (Lonza), and quantitated using Image Studio Lite. Each time course and each mutant assay were repeated three times and estimated errors are given as standard deviations from the mean.
In vivo integration reactions.
Intermolecular recombination in E. coli was measured by transformation of a suicide R6kγ plasmid containing attP (pGV2345 for wild-type attP) into E. coli strain BW25113 containing an attB site in single copy on an F’-episome and an integrase expression plasmid as described [26]. Activity was defined as the transformation efficiency of R6kγ-attP, normalized by the transformation efficiency of pCDFSK, a compatible plasmid carrying a streptomycin resistance marker that was present as an internal control. Agarose gel analysis revealed that many mutants in the library contained plasmid dimers and varying amounts of supercoiled plasmid, complicating quantitative interpretation of the integration activity in terms of transformation efficiency. Activities were therefore scored as + (< 10% of wild-type), ++ (10–60%), or +++ (>60%), based on the average of two independent experiments.
Competition DNA-binding assays.
A competition assay was used to identify weak attPbinding mutants. Binding reactions of 20 μL in EMSA binding buffer contained < 1 nM wildtype attP fragment that was 32P end-labeled as described above for EMSAs, 16 nM Int, and 32 nM mutant attP site (120 bp) that was prepared by PCR amplification of the corresponding mutant plasmid. Preparation of mutant attP sites by PCR was carried out as described above for attB fragments. Binding reactions were incubated at 32°C for 30 min and separated by native PAGE. Electrophoresis, gel scanning, and gel quantitation were all performed as described above for EMSA experiments. Competition binding experiments were carried out 4 or 5 times for each mutant and the results expressed as the average value of 1-fs, where fs = fraction shifted = [Int•DNA]/([Int•DNA] + [DNA]). The value of 1-fs will be high for mutants that are bound with high affinity by Int, since the mutant attP sites will compete effectively for Int binding. Weak binding mutants will have low values of 1-fs, since competition is less effective.
Accession numbers
Protein Data Bank ID: 6DNW
Supplementary Material
Highlights:
Serine integrases are important tools in biotechnology
Listeria innocua Int binds to the attP site with Kd = 7 nM
Only 20 of the 50 attP residues are sensitive to substitution
Specificity is provided by the zinc ribbon and linker regions of Int
This work will facilitate improvement of serine integrase systems
Acknowledgements
This work was supported by the National Institutes of Health [GM108751 to G.V.]. The Advanced Light Source is supported by the Director, Office of Science, Office of Basic Energy Sciences, of the U.S. Department of Energy [contract DE-AC02–05CH11231].
Glossary
- Site-specific recombination
genetic recombination involving specific sequences recognized by site-specific recombinase proteins; DNA strands are exchanged without removal or addition of nucleotides
- attP
integrase binding site found in the phage genome
- attB
integrase binding site and integration site in the host genome
- attL, attR
hybrid sites resulting from integration between attP and attB
- crossover site/sequence
DNA residues where cleavage and strand exchange occur
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Smith MCM, Phage-encoded Serine Integrases and Other Large Serine Recombinases, Microbiology Spectrum. 3 (2015). doi: 10.1128/microbiolspec.MDNA30059-2014. [DOI] [PubMed] [Google Scholar]
- [2].Stark WM, The Serine Recombinases, Microbiology Spectrum. 2 (2014). doi: 10.1128/microbiolspec.MDNA3-0046-2014. [DOI] [PubMed] [Google Scholar]
- [3].Meinke G, Bohm A, Hauber J, Pisabarro MT, Buchholz F, Cre Recombinase and Other Tyrosine Recombinases, Chem. Rev 116 (2016) 12785–12820. doi: 10.1021/acs.chemrev.6b00077. [DOI] [PubMed] [Google Scholar]
- [4].Jayaram M, Ma C-H, Kachroo AH, Rowley PA, Guga P, Fan H-F, Voziyanov Y, An Overview of Tyrosine Site-specific Recombination: From an Flp Perspective, Microbiology Spectrum. 3 (2015). doi: 10.1128/microbiolspec.MDNA3-0021-2014. [DOI] [PubMed] [Google Scholar]
- [5].Grindley N, Whiteson K, Rice P, Mechanisms of site-specific recombination, Annu Rev Biochem. 75 (2006) 567–605. [DOI] [PubMed] [Google Scholar]
- [6].Merrick CA, Zhao J, Rosser SJ, Serine Integrases: Advancing Synthetic Biology, ACS Synth. Biol 7 (2018) 299–310. doi: 10.1021/acssynbio.7b00308. [DOI] [PubMed] [Google Scholar]
- [7].Stark WM, Making serine integrases work for us, Current Opinion in Microbiology. 38 (2017) 130–136. doi: 10.1016/j.mib.2017.04.006. [DOI] [PubMed] [Google Scholar]
- [8].Fogg PCM, Colloms S, Rosser S, Stark M, Smith MCM, New Applications for Phage Integrases, Journal of Molecular Biology. 426 (2014) 2703–2716. doi: 10.1016/j.jmb.2014.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Smith MCM, Thorpe HM, Diversity in the serine recombinases, Mol Microbiol. 44 (2002) 299–307. [DOI] [PubMed] [Google Scholar]
- [10].Groth AC, Calos MP, Phage integrases: biology and applications, J Mol Biol. 335 (2004) 667–78. [DOI] [PubMed] [Google Scholar]
- [11].Landy A, The λ Integrase Site-specific Recombination Pathway, Microbiology Spectrum. 3 (2015). doi: 10.1128/microbiolspec.MDNA3-0051-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Keravala A, Groth AC, Jarrahian S, Thyagarajan B, Hoyt JJ, Kirby PJ, Calos MP, A diversity of serine phage integrases mediate site-specific recombination in mammalian cells, Mol Genet Genomics. 276 (2006) 135–46. [DOI] [PubMed] [Google Scholar]
- [13].Thomason LC, Calendar R, Ow DW, Gene insertion and replacement in Schizosaccharomyces pombe mediated by the Streptomyces bacteriophage phiC31 sitespecific recombination system, Mol Genet Genomics. 265 (2001) 1031–8. [DOI] [PubMed] [Google Scholar]
- [14].Duyne GDV, Cre Recombinase , Microbiology Spectrum. 3 (2015). doi: 10.1128/microbiolspec.MDNA3-0014-2014. [DOI] [PubMed] [Google Scholar]
- [15].Smith MCM, Brown WRA, McEwan AR, Rowley PA, Site-specific recombination by phiC31 integrase and other large serine recombinases, Biochem Soc Trans. 38 (2010) 388–94. [DOI] [PubMed] [Google Scholar]
- [16].Brown WRA, Lee NCO, Xu Z, Smith MCM, Serine recombinases as tools for genome engineering, Methods. 53 (2011) 372–9. [DOI] [PubMed] [Google Scholar]
- [17].Calos MP, The phiC31 integrase system for gene therapy, Curr Gene Ther. 6 (2006) 633–45. [DOI] [PubMed] [Google Scholar]
- [18].Bonnet J, Subsoontorn P, Endy D, Rewritable digital data storage in live cells via engineered control of recombination directionality, PNAS. 109 (2012) 8884–8889. doi: 10.1073/pnas.1202344109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Siuti P, Yazbek J, Lu TK, Synthetic circuits integrating logic and memory in living cells, Nature Biotechnology. 31 (2013) 448–452. doi: 10.1038/nbt.2510. [DOI] [PubMed] [Google Scholar]
- [20].Sclimenti CR, Thyagarajan B, Calos MP, Directed evolution of a recombinase for improved genomic integration at a native human sequence, Nucleic Acids Res. 29 (2001) 5044–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Chalberg TW, Portlock JL, Olivares EC, Thyagarajan B, Kirby PJ, Hillman RT, Hoelters J, Calos MP, Integration specificity of phage phiC31 integrase in the human genome, J Mol Biol. 357 (2006) 28–48. [DOI] [PubMed] [Google Scholar]
- [22].Xiaolai Lei, Lu Wang, Guoping Zhao, Xiaoming Ding, Site-specificity of serine integrase demonstrated by the attB sequence preference of ɸBT1 integrase, FEBS Letters. 592 (2018) 1389–1399. doi: 10.1002/1873-3468.13023. [DOI] [PubMed] [Google Scholar]
- [23].Singh S, Ghosh P, Hatfull GF, Attachment site selection and identity in bxb1 serine integrase-mediated site-specific recombination, PLoS Genet. 9 (2013) e1003490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Gupta M, Till R, Smith MCM, Sequences in attB that affect the ability of phiC31 integrase to synapse and to activate DNA cleavage, Nucleic Acids Res. 35 (2007) 3407–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Rutherford K, Yuan P, Perry K, Sharp R, Van Duyne G, Attachment site recognition and regulation of directionality by the serine integrases, Nuc. Acids. Res. 41 (2013) 8341–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Gupta K, Sharp R, Yuan JB, Li H, Van Duyne GD, Coiled-coil interactions mediate serine integrase directionality, Nucleic Acids Res. 45 (2017) 7339–7353. doi: 10.1093/nar/gkx474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Rowley PA, Smith MCA, Younger E, Smith MCM, A motif in the C-terminal domain of phiC31 integrase controls the directionality of recombination, Nucleic Acids Res. 36 (2008) 3879–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Mandali S, Dhar G, Avliyakulov NK, Haykinson MJ, Johnson RC, The site-specific integration reaction of Listeria phage A118 integrase, a serine recombinase, Mob DNA. 4 (2013) 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Mandali S, Gupta K, Dawson AR, Duyne GDV, Johnson RC, Control of Recombination Directionality by the Listeria Phage A118 Protein Gp44 and the CoiledCoil Motif of Its Serine Integrase, J. Bacteriol. 199 (2017) e00019–17. doi: 10.1128/JB.00019-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Ghosh P, Pannunzio NR, Hatfull GF, Synapsis in phage Bxb1 integration: selection mechanism for the correct pair of recombination sites, J Mol Biol. 349 (2005) 331–48. [DOI] [PubMed] [Google Scholar]
- [31].McEwan AR, Raab A, Kelly SM, Feldmann J, Smith MCM, Zinc is essential for highaffinity DNA binding and recombinase activity of {varphi}C31 integrase, Nucleic Acids Res. (2011). http://view.ncbi.nlm.nih.gov/pubmed/21507889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].McEwan AR, Rowley PA, Smith MCM, DNA binding and synapsis by the large Cterminal domain of phiC31 integrase, Nucleic Acids Res. 37 (2009) 4764–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Van Duyne GD, Rutherford K, Large serine recombinase domain structure and attachment site binding, Crit. Rev. Biochem. Mol. Biol 48 (2013) 476–491. doi: 10.3109/10409238.2013.831807. [DOI] [PubMed] [Google Scholar]
- [34].Thyagarajan B, Olivares EC, Hollis RP, Ginsburg DS, Calos MP, Site-Specific Genomic Integration in Mammalian Cells Mediated by Phage φC31 Integrase, Mol. Cell. Biol 21 (2001) 3926–3934. doi: 10.1128/MCB.21.12.3926-3934.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Ghosh P, Kim AI, Hatfull GF, The orientation of mycobacteriophage Bxb1 integration is solely dependent on the central dinucleotide of attP and attB, Mol Cell. 12 (2003) 1101–11. [DOI] [PubMed] [Google Scholar]
- [36].Smith MCA, Till R, Smith MCM, Switching the polarity of a bacteriophage integration system, Mol Microbiol. 51 (2004) 1719–28. [DOI] [PubMed] [Google Scholar]
- [37].Ghosh P, Wasil LR, Hatfull GF, Control of phage Bxb1 excision by a novel recombination directionality factor, PLoS Biol. 4 (2006) e186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Khaleel T, Younger E, McEwan AR, Varghese AS, Smith MCM, A phage protein that binds φC31 integrase to switch its directionality, Mol Microbiol. (2011). http://view.ncbi.nlm.nih.gov/pubmed/21564337. [DOI] [PubMed] [Google Scholar]
- [39].Zhang L, Zhu B, Dai R, Zhao G, Ding X, Control of Directionality in Streptomyces Phage φBT1 Integrase-Mediated Site-Specific Recombination, PLoS ONE. 8 (2013) e80434. doi: 10.1371/journal.pone.0080434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Bogdanove AJ, Bohm A, Miller JC, Morgan RD, Stoddard BL, Engineering altered protein–DNA recognition specificity, Nucleic Acids Res. (n.d.) doi: 10.1093/nar/gky289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Akopian A, He J, Boocock MR, Stark WM, Chimeric recombinases with designed DNA sequence recognition, Proc Natl Acad Sci U S A. 100 (2003) 8688–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Gill S, von Hippel P, Calculation of protein extinction coefficients from amino acid sequence data, Anal Biochem. 182 (1989) 319–26. [DOI] [PubMed] [Google Scholar]
- [43].Kabsch W, Integration, scaling, space-group assignment and post-refinement, Acta Cryst D, Acta Cryst Sect D, Acta Crystallogr D, Acta Crystallogr Sect D, Acta Crystallogr D Biol Crystallogr, Acta Crystallogr Sect D Biol Crystallogr. 66 (2010) 133–144. doi: 10.1107/S0907444909047374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ, Phaser crystallographic software, J Appl Crystallogr. 40 (2007) 658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL, Crystallography & NMR system: A new software suite for macromolecular structure determination, Acta Crystallogr D Biol Crystallogr. 54 (1998) 905–21. [DOI] [PubMed] [Google Scholar]
- [46].Emsley P, Cowtan K, Coot: model-building tools for molecular graphics, Acta Crystallogr D Biol Crystallogr. 60 (2004) 2126–32. [DOI] [PubMed] [Google Scholar]
- [47].Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung L-W, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH, PHENIX: a comprehensive Python-based system for macromolecular structure solution, Acta Crystallogr D Biol Crystallogr. 66 (2010) 213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Delano W, The PyMOL Molecular Graphics System, DeLano Scientific, San Carlos, 2002. [Google Scholar]
- [49].Crooks GE, Hon G, Chandonia J-M, Brenner SE, WebLogo: A Sequence Logo Generator, Genome Res. 14 (2004) 1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.