Abstract
Fitness cost is a common phenomenon in rice blast disease-resistance breeding. MiR396 is a highly conserved microRNA (miRNA) family targeting Growth Regulating Factor (OsGRF) genes. Mutation at the target site of miR396 in certain OsGRF gene or blocking miR396 expression leads to increased grain yield. Here we demonstrated that fitness cost can be trade-off in miR396-OsGRFs module via balancing growth and immunity against the blast fungus. The accumulation of miR396 isoforms was significantly increased in a susceptible accession, but fluctuated in a resistant accession upon infection of Magnaporthe oryzae. The transgenic lines over-expressing different miR396 isoforms were highly susceptible to M. oryzae. In contrast, overexpressing target mimicry of miR396 to block its function led to enhanced resistance to M. oryzae in addition to improved yield traits. Moreover, transgenic plants overexpressing OsGRF6, OsGRF7, OsGRF8, and OsGRF9 exhibited enhanced resistance to M. oryzae, but showed different alteration of growth. While overexpression of OsGRF7 led to defects in growth, overexpression of OsGRF6, OsGRF8, and OsGRF9 resulted in better or no significant change of yield traits. Collectively, our results indicate that miR396 negatively regulates rice blast disease- resistance via suppressing multiple OsGRFs, which in turn differentially control growth and yield. Therefore, miR396-OsGRFs could be a potential module to demolish fitness cost in rice blast disease-resistance breeding.
Keywords: disease resistance, fitness cost, miR396, Oryza sativa, OsGRF, rice blast disease
Introduction
Plants are often targeted by pathogens such as bacteria, fungi, and oomycetes, and as a part of evolution, they developed complex network comprising pathogen-associated molecular pattern- (PAMP)-triggered immunity (PTI) and Effector-triggered immunity (ETI), to counteract the pathogens (Ramirez-Prado et al., 2018). Accumulating evidence indicates that plant microRNAs play important roles in both PTI and ETI responses against pathogens (Seo et al., 2013; Deng et al., 2018). For example, MiR393 regulates host immunity against Pseudomonas syringae DC3000 by suppressing auxin signaling through targeting the auxin receptors such as TIR1, AFB2, and AFB3 (Navarro et al., 2016). Following studies identified miR160 as a positive regulator, whereas, miR825, miR398b, and miR773 as negative regulators of immunity against bacterial infection in Arabidopsis (Fahlgren et al., 2007; Jagadeeswaran et al., 2009; Li et al., 2010). In tomato, miR482 regulates host defense by targeting coiled-coil-nucleotide-binding leucine-rich-repeat (CC-NB-LRR)-encoding transcripts and triggers the production of secondary siRNAs via RDR6, which in turn targets other mRNAs of defense related genes (Shivaprasad et al., 2012). In tobacco, nta-miR6019 and nta-miR6020 targets the immune receptor gene N that confers resistance to tobacco mosaic virus (Li et al., 2012). In apple, Md-miRLn11 regulates NBS-LRR gene expression upon apple leaf spot fungal infection (Ma et al., 2014).
Rice is the most widely cultivated food crop in the world and its production is immensely affected by rice blast disease caused by M. oryzae (Baldrich and San Segundo, 2016). Similar to Arabidopsis, rice also involves two layered immune system (PTI and ETI) against M. oryzae (Chen and Ronald, 2011). In rice, PTI is mediated by Pattern Recognition Receptors (PRRs) such as CEBiP, LYP4, and LYP6 by effectively chitin recognition, and the second layer of immunity is mostly modulated by nucleotide binding site/ Leucine rich repeat (NBS-LRR) proteins (Chen and Ronald, 2011). So far, 102 rice blast resistance (R) genes have been identified, out of which 28 genes were functionally characterized (Li W.T. et al., 2017). Increasing evidence shows that miRNAs modulate responses to M. oryzae infection by effectively regulating their target genes (Campo et al., 2013; Li et al., 2014; Baldrich and San Segundo, 2016). For example, Osa-miR7695 is the first miRNA that was identified to promote resistance to M. oryzae by negatively regulating its target transcript, OsNramp6 (Campo et al., 2013). In addition, many new miRNAs, conserved and non-conserved miRNAs were identified to be differentially responsive to M. oryzae or its elicitor (Li et al., 2014; Baldrich et al., 2015). Deeper investigation focuses on functionally characterization of each miRNA and its roles in rice blast-disease resistance are disclosed one after another. In the past few years, several groups have identified that miR164a, miR169a, miR319b, and miR444b.2 negatively regulate, whereas, miR160a, miR166k-166h, miR398b, and miR7695 positively regulate rice immunity against the rice blast fungus M. oryzae (Campo et al., 2013; Li et al., 2014; Li Y. et al., 2017; Xiao et al., 2017; Salvador-Guirao et al., 2018; Wang et al., 2018; Zhang et al., 2018). However, the roles of other blast fungus-responsive miRNAs remain elusive.
In our previous study, we identified a number of miRNAs that were differentially regulated upon M. oryzae infection in susceptible and resistant rice accessions (Li et al., 2014). Among them, miR396 family members were differentially expressed. MiR396d/e-5p and miR396e-3p were induced in the susceptible accession LTH at 12 and 24 h post inoculation (hpi), respectively; whereas, found no change in the resistant accession IRBLkm-Ts. However, miR396c-5p was increased in both LTH and IRBLkm-Ts. Therefore, miR396 might be involved in regulation of rice immunity against M. oryzae. However, an in-depth study is essential to elucidate the role of miR396 family members against rice blast fungus due to its multiple isoforms targeting the same group of genes encoding Growth Regulating Factors (GRFs) (Liu H.H. et al., 2014).
Growth Regulating Factors are plant-specific transcription factors defined by the presence of highly conserved WRC and QLQ protein domains that are recognized for their roles in stem, root and leaf development, flower and seed formation, and also maintain growth under adverse environmental conditions (Omidbakhshfard et al., 2015). In rice, the GRF transcription factor family comprises 12 members whose expression are epigenetically controlled by miR396 to regulate multiple biological processes (Liu H.H. et al., 2014). For example, miR396-OsGRF components are involved in floral organogenesis (Liu H.H. et al., 2014), regulation of grain size and yield (Che et al., 2016; Duan et al., 2016; Gao et al., 2016; Li et al., 2016) and plant architecture (Tang et al., 2018). Recently, evidence has also indicated the role of miR396 during pathogen infection. miR396 expression was significantly reduced in wheat line JD8 (susceptible) and JD8-Pm30 (resistant line) when infected by Erysiphe graminis f. sp. tritici (Xin et al., 2010), and in wheat accession Shan 4445 (susceptible) when infected by Blumeria graminis (Wu et al., 2015). Overexpression of Sp-miR396a-5p in tobacco leads to enhanced susceptibility to Phytophthora nicotianae infection (Chen et al., 2015). More recently, miR396 was found to regulate defense against hemibiotrophic and necrotrophic fungal pathogens in Arabidopsis (Soto-Suarez et al., 2017). However, the underlying mechanism is largely unclear.
In order to elucidate the role of miR396 in rice immunity against the blast fungus, we analyzed the expression of miR396 isoforms and their target genes in LTH (susceptible) and IRBLkm-Ts (resistant) accessions. Further, we examined the blast disease phenotypes in transgenic lines over-expressing four miR396 isoforms and their target mimicry, respectively. Based on target gene expression in transgenic lines, resistant and susceptible accessions, we selected OsGRF6, OsGRF7, OsGRF8, and OsGRF9 for further studies. Overexpression of these target genes resulted in enhanced resistance to rice blast fungal infection. In addition, miR396 target mimic and OsGRF6 overexpressing transgenic lines showed improved yield-traits, in contrast to the transgenic lines overexpressing OsGRF7 showing growth defects. Overall, our data illustrate that miR396-GRFs module regulates rice immunity against blast fungus, which can be combined with improvement of yield-traits. Therefore, miR396-GRFs module is highly valuable in disease-resistance and high yield-breeding programs.
Materials and Methods
Plant Materials and Growth Conditions
The plant materials used in this study include LTH (M. oryzae susceptible) and IRBLkm-Ts (M. oryzae resistant) lines. The miR396 overexpression and STTM396 (short tandem target mimic of miR396) transgenic lines were obtained from a previous study (Li et al., 2016) (under Nipponbare, a Japonica accession); and miR396 target mimic (MIM396) and OsGRF overexpression transgenic lines were also obtained from a previous study (Gao et al., 2016) (under Yuetai-B, an Indica accession). The rice plants were maintained in the field of Sichuan Agricultural University, Wenjiang, Chengdu. N. benthamiana plants were maintained at 22°C with 16/8 h of day/night light in a growth chamber and used for agro-infiltration experiments.
Plasmid Construction and Genetic Transformation
MiR396 over expression plasmid constructs were provided by Li et al. (2016). Artificial target mimicry sequences of miR396d were inserted into the IPS1 to replace the miR399 target site with primers osa-miR396d – MIMIC FP and osa-miR396d – MIMIC RP (Supplementary Table S3) as described previously (Franco-Zorrilla et al., 2007; Li Y. et al., 2017) and cloned into BamHI–BglII sites of binary vector 35S-pCAMBIA1300, resulting in over-expressing construct p35S:MIM396d. Reporter plasmids were constructed by inserting the artificial sequences of miR396 target gene site with specific primers (Supplementary Table S3) at the beginning of eYFP codon sequences and cloned into the KpnI site of binary vector 35S-pCAMBIA1300. The constructed plasmids were used for Agrobacterium- (GV3101) mediated transient expression assay in N. benthamiana.
Pathogen Growth and Infection
Four M. oryzae strains, Guy11, Zhong1, NC10, and eGFP-tagged Zhong8- 10-14 (GZ8) were used in this study. M. oryzae strains were cultured in complete medium (10 g/L D-glucose, 2 g/L Peptone, 1 g/L Yeast Extract, 1 g/L Casamino Acids, 0.1% (v/v) Vitamin Solution, 5% (v/v) Nitrate Salts (120 g/L NaNO3, 10.4 g/L KCL, 5.2 g/L MgSO4, 30.4 g/L KH2PO4), 0.1% (v/v) Trace Elements (22 g/L ZnSO4⋅7H2O, 11 g/L H3BO3, 5 g/L MnCl2⋅4H2O, 5 g/L FeSO4⋅7H2O, 1.7 g/L CoCl2⋅6H2O, 1.6 g/L CuSO4⋅5H2O, 1.5 g/L NaMoO4⋅2H2O, 50 g/L Na4EDTA), 15 g/L agar, pH 6.5) at 28°C with 12-h/12-h light/dark cycles for sporulation. After 2 weeks, spores were collected and the inoculum concentration was adjusted to 1 × 105 spores mL-1 for assays. Three-leaf-stage seedlings were used for spray inoculation using 1 × 105 spores mL-1 and the disease phenotypes on leaf two were recorded after 5 days post inoculation (dpi). For punch inoculation method (Park et al., 2012), 5 μL of spore suspension (1 × 105 spores mL-1) was added at two spots of each leaf, kept in a culture dish containing 0.1% 6-Benzylaminopurine (6-BA). Lesion length was measured after 5 dpi. Relative fungal mass was calculated using DNA concentration of M. oryzae Pot 2 against the rice genomic ubiquitin DNA level by qPCR (Park et al., 2012; Li W.T. et al., 2017. The infection process of fungus and examination of H2O2 were performed according to Xiao et al. (2003) and Kankanala et al. (2007)), respectively. All quantification analyses of H2O2 area were conducted in Photoshop according to a previous report (Li W.T. et al., 2017).
RNA Isolation and Quantitative RT-PCR
Total RNA was extracted from collected samples using TRIzol reagent (Invitrogen). RNA quality and quantity were determined by spectrophotometer and using electrophoresis. Reverse transcription to cDNA was done using the SuperScript first-strand synthesis kit (Invitrogen). To analyze the expression of miRNA, stem-loop pulse RT-qPCR (Varkonyi-Gasic et al., 2007) was performed using the designed stem-loop containing RT and forward primer containing the 5′ end of mature miRNA (forward and reverse) designed according to Varkonyi-Gasic et al. (2007). For internal reference, primers specific to U6 snRNA (Turner et al., 2013) was used. The rice ubiquitin (UBQ) gene was used as an internal reference for normalizing target gene expression.
Transient Expression Assay in N. benthamiana
Agrobacterium strain GV3101 containing the individual expression constructs in the binary vector pCAMBIA1300 was incubated at 28°C overnight in LB media containing specific antibiotics at a 250 r/min shaking incubator. The bacteria were collected at 3000 rpm for 5 min and re-suspended in an MMA buffer (10 mM MES, 10 mM MgCl2, and 100 mM AS). The re-suspended culture was infiltrated into the leaves of N. benthamiana for transient expression assay. After 36 hpi, the infiltrated leaves were observed for image acquisition using a NikonA1 Confocal Laser Scanning Microscope (Nikon Instruments, Inc., Chengdu, China) as previously described (Huang et al., 2014). Western blotting analyses were performed following a previous protocol (Chen et al., 2008). Samples were also collected for RNA isolation and RT-qPCR as described above.
Results
Differential Responses of miR396 Isoforms to M. oryzae in Susceptible and Resistant Accession
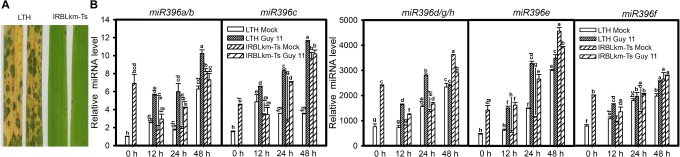
MiR396 is a conserved and highly abundant miRNA family reported in most plants including rice and Arabidopsis. The rice genome contains eight loci encoding miR396 (with 5 mature isoforms) distributed on 3 different chromosomes: miR396a, miR396b, miR396c, miR396d, miR396e, miR396f, miR396g, and miR396h (miRBase release 22, Supplementary Figure S1). In order to elucidate the differential responses of miR396 isoforms to M. oryzae infection, we analyzed their expression in the susceptible line LTH and the monogenic resistant line IRBLkm-Ts that contains the resistance (R) gene Pi-km and exhibits high resistance to M. oryzae isolates carrying AVR-Pikm (Li et al., 2014). Initially, we confirmed the blast disease phenotype of LTH and IRBLkm-Ts by inoculating with Guy11 spores on three-leaf-stage seedlings as described in the Section “Materials and Methods” (Figure 1A). According to the small RNA deep sequencing data from our previous studies, the accumulation of miR396d/e is mostly abundant, followed by miR396c, whereas miR396a and miR396f is very less (Li et al., 2014). In the present study, the accumulation of miR396d, miR396e, and miR396f were increased approximately 3800, 4500, and 3000 folds, respectively, whereas miR396a/b and miR396c expression levels were increased 10 to 12 folds (Figure 1B). Therefore, the data confirmed that the accumulation of different miR396 isoforms was different and are consistent with earlier deep sequencing results. The expression of miR396a/b and miR396c (Figure 1B) were increased at all the three time points tested in LTH, whereas increased only at one or two time points in IRBLkm-Ts. Similarly, miR396d/g/h, miR396e, and miR396f were highly increased at all the three time points in LTH, whereas slightly increased only at 12 and 24 hpi in IRBLkm-Ts. Overall, the data indicate that miR396 isoforms are differentially responsive to M. oryzae in rice.
FIGURE 1.
Magnaporthe oryzae infection induces differential accumulation of miR396 isoforms in susceptible and resistant rice accessions. (A) Representative leaf section of LTH (susceptible) and IRBLkm-Ts (resistant) showing the blast disease phenotype. (B) Accumulation of the indicated miR396 isoforms in LTH and IRBLkm-Ts upon M. oryzae (or mock inoculated) infection. RNA samples were extracted from leaves collected at different time points and the expression of the indicated miR396 isoforms were performed using Stem-loop RT-qPCR. SnRNA U6 served as the internal reference. Error bars indicate SD from three technical replicates. The letters above the bars indicate significant differences at a P-value < 0.01, as determined by a one-way ANOVA followed by post hoc Tukey HSD analysis. The experiment was repeated twice with similar results.
Overexpression of miR396 Enhances Susceptibility to M. oryzae
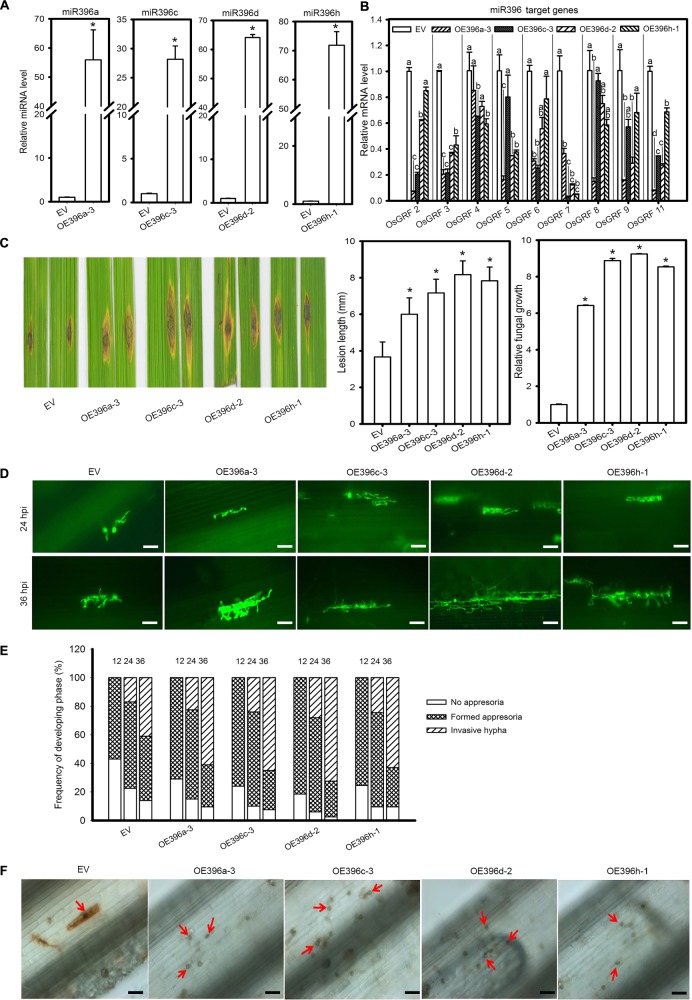
Based on the above results we speculated that miR396 might negatively regulate rice immunity against M. oryzae. Because all miR396 isoforms target the same group of OsGRF genes (Supplementary Figures S1B, S2), we tested four miR396 isoforms in response to the blast fungus. Therefore, we obtained transgenic rice plants overexpressing miR396a, miR396c, miR396d and miR396h (OE396) under Nipponbare (NPB) background. Then, the transgenic lines were subjected to rice blast disease assay. Stem-loop RT-qPCR confirmed the increased accumulation of miR396 isoforms (Figure 2A). Correspondingly, the expression of OsGRFs were significantly suppressed in the OE396 lines (Figure 2B). Rice blast disease assay was conducted by punch-inoculation with M. oryzae strain Zhong1 using detached leaves. Intriguingly, all the OE396 lines exhibited enhanced susceptibility forming lesions larger than control plants (Figure 2C). The lesion lengths were increased up to 1.64, 1.95, 2.22, and 2.13 folds of control plants in OE396a, OE396c, OE396d, and OE396h, respectively (Figure 2C). Fungal biomass quantification also showed that the OE396 lines supported significantly more fungal growth than control plants (Figure 2C).
FIGURE 2.
Overexpression of four miR396 isoforms resulted in enhanced susceptibility to M. oryzae. (A) Accumulation of the indicated four miR396 isoforms in the respective transgenic lines. (B) Reduction of OsGRF genes in the indicated transgenic lines. (C) Blast disease assay on the indicated lines. Punch inoculation of M. oryzae strain Zhong1 on 4–5 week old leaves from wild type EV (NPB) and the indicated transgenic lines overexpressing four miR398 isoforms. Disease severity was recorded and evaluated at 5 days post inoculation. Relative fungal biomass is determined by examining the expression level of M. oryzae Pot2 gene against OsUbiquitin DNA level. (D) Representative epi-fluorescent images of sheath cells from EV and the indicated transgenic lines infected by eGFP-tagged blast isolate GZ8. Bars = 100 μm. (E) Quantification analysis on the progress of fungal infection at 12, 24, and 48 hpi. Around 200 conidia in each line were analyzed. Similar results were obtained in at least two independent experiments. (F) DAB staining shows H2O2 accumulation at the infection sites of the indicated lines at 2 days post inoculation (dpi). Bars = 100 μm. Arrows indicate appressoria. In (A–C), error bars indicate SD (n = 3). The letters above the bars indicate significant differences at a P-value < 0.01, as determined by a one-way ANOVA followed by post hoc Tukey HSD analysis. The asterisk above the bars indicate significant differences between EV and the indicated transgenic lines at a P-value < 0.01, as determined by Student’s t-test.
To understand how OE396 lines supported more fungal growth, we examined the infection process of the eGFP-tagged strain Zhong10-8-14 (GZ8) on sheath cells under epifluorescence microscopy. At 12 hpi, appressoria were formed on sheaths of control and transgenic plants. At 24 hpi, most spores formed invasive hyphae on both control and OE396 lines. At 36 hpi, 65–70% of spores formed invasive hyphae on OE396 lines; by contrast, only ∼50% formed on control lines (Figures 2D,E). These results indicate that overexpression of the four miR396 isoforms highly facilitates the growth of M. oryzae.
One typical defense response to pathogen infection is the production of H2O2 that can be demonstrated by 3, 3′-diaminobenzidine (DAB)-staining. Here, DAB-staining of the M. oryzae inoculated leaf sheath showed that the control plants obviously produced H2O2 in the inoculated sheath at 48 hpi (determined by the staining at the infection site), whereas H2O2 was hardly detected in the OE396 lines (Figure 2F). The data indicate that overexpression of miR396 may suppress rice defense response, facilitating the growth of M. oryzae. Altogether, overexpression of miR396 leads to enhanced susceptibility to M. oryzae.
Transcriptional Regulation of OsGRFs by miR396 Isoforms
The potentiality of a miRNA is anticipated on how strong it suppresses target genes. Because overexpression of miR396 leads to suppression of target genes, we speculated to identify the preferential target genes whose expression is more significantly reduced in the OE396 lines. Apart from OsGRF1, OsGRF10, and OsGRF12 that we could not detect by RT-qPCR, which might be due to their panicle-specific expression, the expression of the other nine GRF genes, including OsGRF2, OsGRF3, OsGRF4, OsGRF5, OsGRF6, OsGRF7, OsGRF8, OsGRF9, and OsGRF11, were significantly suppressed in at least one of the four OE396 lines (Figure 2B). However, the levels of suppression were different among transgenic lines expressing different miR396 isoforms (Figure 2B), indicating preferential targeting of different miR396 isoforms toward different OsGRF target sites. While miR396a seems to be the most potent isoform that could significantly suppress all detected OsGRFs except OsGRF4, miR396c, miR396d and miR396h could significantly suppress six of the nine detected OsGRFs (Figure 2B).
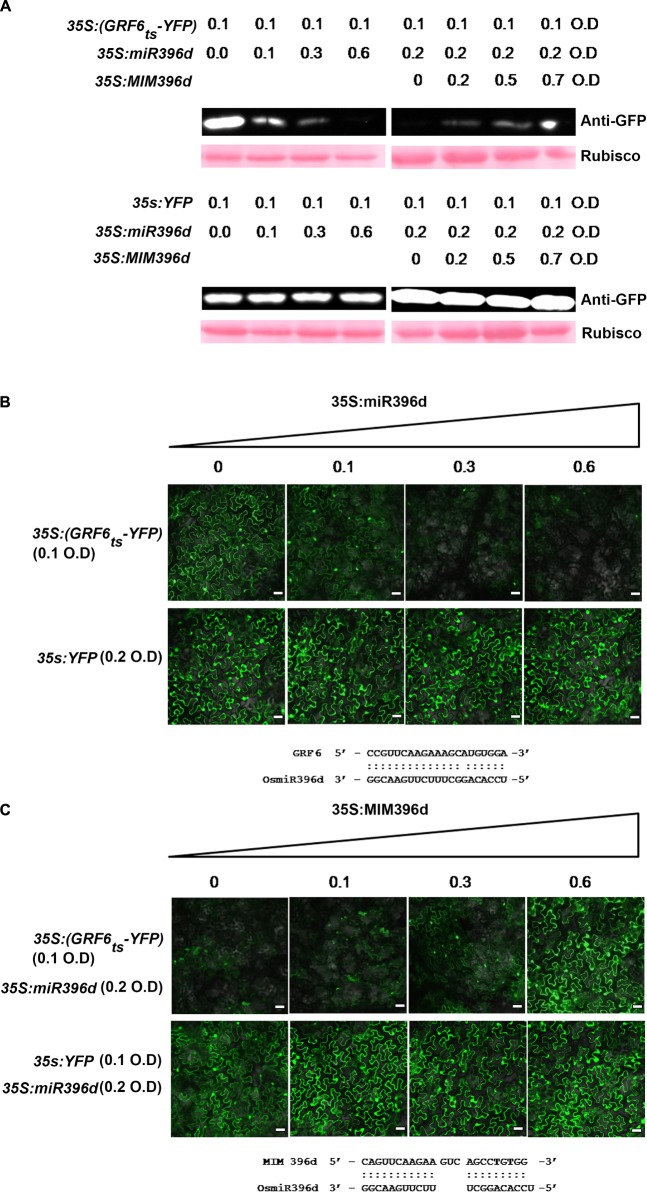
To verify the preferential targeting of OsGRFs by different miR396 isoforms, we performed a Yellow Fluorescent Protein (YFP)-based reporter assay, which was transiently expressed in Nicotiana benthamiana as described in a previous report (Li Y. et al., 2017). Briefly, we constructed a vector expressing YFP containing the target site of OsGRF6 at the beginning of eYFP coding sequences (35S:GRF6ts-YFP). The Agrobacteria containing 35S:GRF6ts-YFP was infiltrated separately or in combination with miR396d in N. benthamiana and the protein levels were analyzed by western blotting and by examining the intensity of YFP in the Nikon A1 Confocal Laser Scanning Microscope (Nikon Instruments, Inc., Chengdu, China). When 35S:GRF6ts-YFP was expressed alone, it was highly accumulated with intensive signals of YFP (Figures 3A,B). However, when co-expressed with miR396d, the expression was gradually decreased with increased concentration of Agrobacteria harboring miR396d. Conversely, there was no change in the protein concentration or intensity of YFP when the YFP vector without target site or with mutant OsGRF6 site was expressed at the same conditions (Figures 3A,B). To further validate the targeting of miR396d on 35S:GRF6ts-YFP, target mimicry of miR396d (MIM396d) was constructed to trap miR396d in order to reduce its efficiency to bind 35S:GRF6ts-YFP. When 35S:GRF6ts-YFP was co-expressed along with miR396d and MIM396d, the protein expression was gradually increased with the increased concentration of Agrobacteria harboring MIM396d (Figures 3A,C). The results validated the reporter system.
FIGURE 3.
MiR396 represses the expression of its target genes at translational level. (A) Western blotting analysis and (B,C) confocal images (bars = 25 μm) show that miR396d suppressed the accumulation of GRF6ts-YFP but did not affect the protein level of YFP control. The indicated 35S:GRF6ts-YFP and YFP-based reporter constructs were transiently expressed alone or co-expressed with miR396d (B) or/and the miR396 target mimicry MIM396d (C) in Nicotiana benthamiana leaves using Agrobacterium-mediated infiltration at the indicated optical density (O.D.) concentration. Protein extracts from the same amount of infiltrated leaves were subjected to Western blot analysis using anti-GFP sera. The Ponceau S stained Rubisco served as loading control (A). The alignments of miR396d with OsGRF6 target sequence (B) and MIM396d with miR396d (C) were listed below the images, respectively.
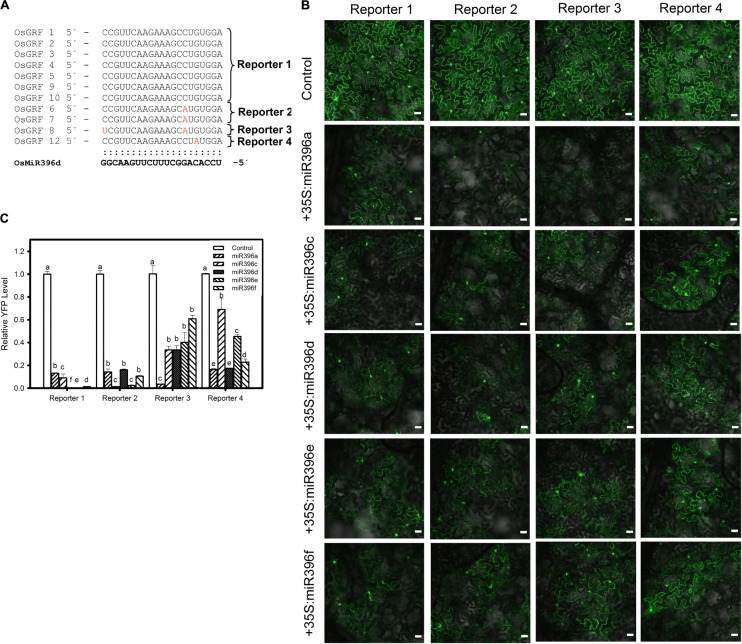
Next, we conducted the similar experiment with different isoforms of miR396 and OsGRFs by analyzing their expression at both translational and transcriptional level. Each of five different plasmids (35S:miR396a, 35S:miR396c, 35S:miR396d, 35S:miR396e, and 35S:miR396f) was co-expressed separately with each of the four reporters (Reporter 1, Reporter 2, Reporter 3, and Reporter 4, Figure 4A) in N. benthamiana. Our data showed that all the miR396 isoforms can suppress different target sites of OsGRFs, however, we could not distinguish the favorite target site based on the YFP intensity (Figure 4B). Therefore, RT-qPCR analysis was further conducted and showed that Reporter 1 and 2, constituting the most number (nine) of target genes, were highly suppressed when compared to reporter 3 and 4 (each constitutes one gene) (Figures 4A,C), indicating that miR396 family members target some sites stronger than the others. This could be due to the location and number of mismatched nucleotides. Altogether, the above data indicate that different miR396 isoforms differentially regulate different OsGRFs at transcriptional level.
FIGURE 4.
Preferential targeting of miR396 isoforms against OsGRFs. (A) Multiple alignment of miR396d target sites in OsGRF’s. The target genes were classified into four categories (Reporter 1, Reporter 2, Reporter 3, and Reporter 4) based on sequence homology of target site. (B) Confocal images show that miR396 isoforms differentially suppresses the protein accumulation of different OsGRF’s reporter genes. The indicated YFP-based reporter constructs were transiently expressed alone or co-expressed with miR396 isoforms in Nicotiana benthamiana. Bars = 25 μm. (C) RNA samples were collected from infiltrated leaves and the transcriptional regulation of OsGRF’s homologs by miR396 isoforms were studied using RT-qPCR. Error bars indicate SD from three technical replicates. The letters above the bars indicate significant differences at a P-value < 0.01, as determined by a one-way ANOVA followed by post hoc Tukey HSD analysis. This experiment was repeated at least three times with similar results.
Overexpression of Target Mimicry of miR396 Enhances Resistance to M. Oryzae and Improves Yield Traits
To gain more insight on the role of miR396 in rice immunity against blast fungus, we assessed the infectivity of M. oryzae against the transgenic plants expressing the target mimicry of miR396 (MIM396) under Yuetai-B (YB) background. The relative expression levels of miR396 isoforms were significantly down-regulated in MIM396 lines (Figure 5A). All the detected target genes, except OsGRF2 and OsGRF11, were significantly increased from 2 to 12 folds in MIM396 lines (Figure 5B). Particularly, OsGRF6 level has increased 7 to 12 folds compared to control plants. We next examined the resistance of MIM396 lines against M. Oryzae (Zhong1). To our expectation, MIM396 lines showed enhanced resistance as indicated by the lesions smaller than those of the control plants (Figure 5C). The lesion length on the MIM396 lines was significantly reduced when compared to control plants (Figure 5C). Consistently, fungal biomass was significantly decreased in MIM396 lines compared to control plants (Figure 5C). Similar results were obtained when the MIM396 lines were infected with the blast isolate NC10 (Supplementary Figure S3). Correspondingly, we examined the rice blast infectivity in transgenic lines over-expressing STTM396 (Nipponbare background) from a previous study (Li et al., 2016). As expected, the STTM396 lines exhibited enhanced resistance to rice blast disease (Supplementary Figure S4). The lesion length was significantly reduced compared to control plants (Supplementary Figure S4). Quantification of fungal biomass confirmed the lesion length results (Supplementary Figure S4).
FIGURE 5.
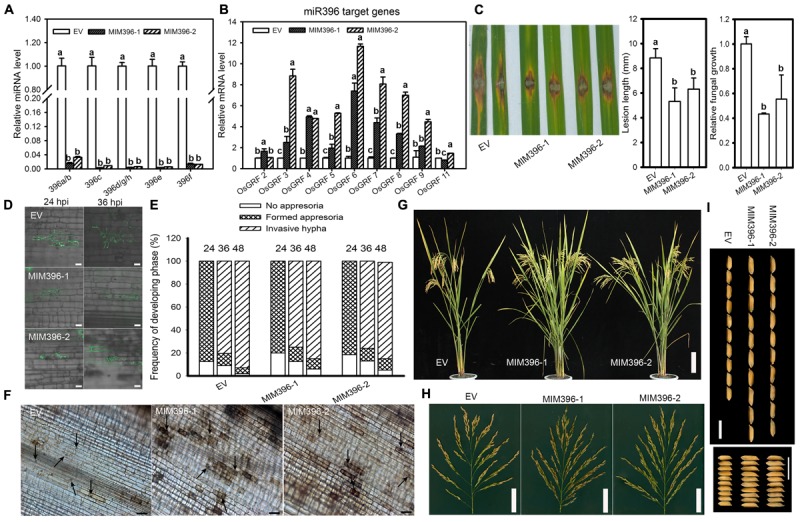
Overexpression of miR396 target mimicry results in enhanced resistance to M. oryzae with improved yield traits. RT-qPCR analyses show the expression of miR396 isoforms (A) and its target genes (B) in control plants (EV, empty vector in YB background) and transgenic lines over-expressing target mimicry of miR396. The accumulation level was normalized to that of the control plants. (C) Punch inoculation of 4–5 week old leaves from EV (YB), MIM396-1 and MIM396-2 rice plants show disease severity of M. oryzae (Zhong1, 1 × 105 spore/ml conc.) at 5 day post inoculation. Relative fungal biomass is determined by examining the expression level of M. oryzae Pot2 gene against OsUbiquitin DNA level. (D) Representative confocal images of sheath cells from EV, MIM396-1 and MIM396-2 infected by eGFP-tagged blast isolate GZ8. Bars = 20 μm. (E) Quantification analysis on the progress of fungal infection at 24, 36, and 48 hpi. Around 200 conidia in each line were analyzed. Similar results were obtained in at least two independent experiments. (F) DAB staining shows H2O2 accumulation at the infection sites of the indicated lines at 2 days post inoculation (dpi). Arrows indicate appressoria. Bars = 100 μm. (G–I) Comparison of gross plant (G), scale bar = 14 cm; and panicle morphologies (H), scale bar = 5 cm; and seed length and seed width (I), scale bar = 10 mm, of EV (YB) and miR396 mimic (MIM396-1 and MIM396-2) plants. In (A–C), error bars indicate SD from three technical replicates. The letters above the bars indicate significant differences at a P-value < 0.01, as determined by a one-way ANOVA followed by post hoc Tukey HSD analysis.
We next examined the infection process of GZ8 on sheath cells of control and MIM396 lines. At 12 hpi, appressoria formation was hardly detected; whereas at 24 hpi most spores formed appressoria on control and MIM396 lines. At 48 hpi, ∼95% of spores formed invasive hyphae on control plants; in contrast, only 80–85% formed on MIM396 lines (Figures 5D,E). In addition, DAB-staining showed that the MIM396 lines produced obviously more H2O2 than the control plants (Figure 5F), which was consistent with the resistance phenotype.
Regarding agronomic traits, the MIM396 lines showed a significant increase in yield-traits compared to control plants, such as the panicle branches, spikelet numbers (Figures 5G,H) and grain size (Figure 5I), which was consistent with a previous report (Gao et al., 2016). To further confirm the growth by field trials, we investigated the agronomic traits in two different experimental locations (Wuhan, ∼N31-E114, and Chengdu, ∼N30-E102, Supplementary Table S2). They showed increased yield traits and were consistent with a previous report (Gao et al., 2016). In total, the above results indicate that blocking miR396 improves both rice blast resistance and yield traits.
Differential Expression of miR396 Target Genes in Susceptible and Resistant Rice Accession
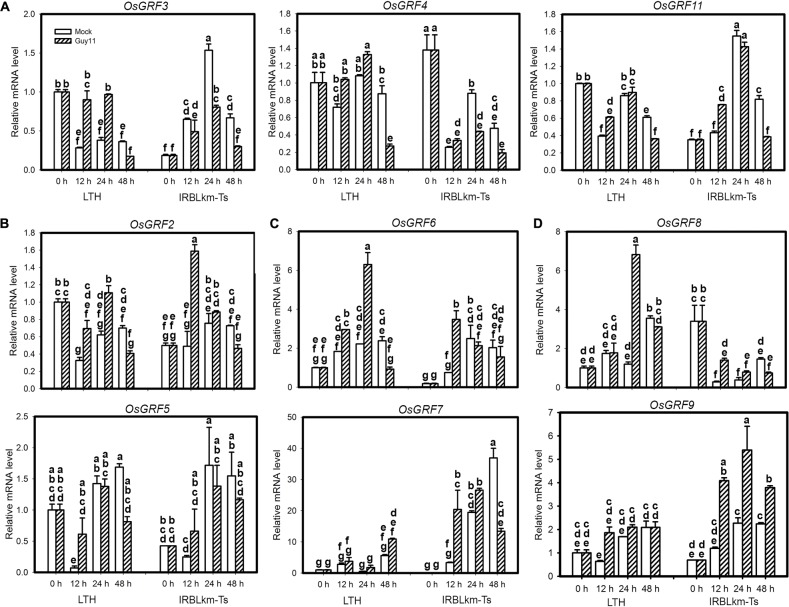
The above data emphasize the role of miR396 target genes in rice blast resistance. However, it seems that all the 12 OsGRFs are regulated by miR396 and it is challenging to identify the one or ones acting in immunity. Therefore, we performed a time course study examining their expression in LTH and IRBLkm-Ts upon M. oryzae infection. Based on their expression patterns, they were classified into four groups (Figure 6). The first group included three genes, namely OsGRF3, OsGRF4 and OsGRF11, their transcription levels were increased in LTH alone at two time points, whereas decreased at least in one time points in IRBLkm-Ts (Figure 6A). The second group contained OsGRF2 and OsGRF5, their expression pattern in LTH was similar to group 1, whereas, in IRBLkm-Ts the expression levels were significantly increased at 12 hpi, and then decreased or no significant change at later time points (Figure 6B). The third group contained OsGRF6 and OsGRF7, their expression levels were increased in both LTH and IRBLkm-Ts, and however, the expression fold change is much higher compared to first and second group (Figure 6C). The fourth group contained OsGRF8 and OsGRF9, the expression levels were mostly stable or increased at one time point in LTH, whereas in IRBLkm-Ts, the expression levels were significantly increased at least in two time points (Figure 6D). Even though, most genes were up-regulated in LTH, we observed differential expression patterns, indicating that different GRFs may act differentially in rice immunity against blast fungus. Genes in group 3 (OsGRF6 and OsGRF7) and group 4 (OsGRF8 and OsGRF9) may positively regulate rice immunity against M. oryzae.
FIGURE 6.
Differential expression of miR396’s target genes in susceptible and resistant accessions upon M. oryzae infection. (A–D) RT-qPCR analyses show the expression of the indicated OsGRF genes in LTH and IRBLkm-Ts at the indicated time points upon M. oryzae infection. Relative mRNA level was normalized to that in mock samples before inoculation (0h). Values are means of three technical replicates. Error bars indicate SD. The letters above the bars indicate significant differences (P < 0.01) as determined by a one-way ANOVA followed by post hoc Tukey HSD analysis.
Overexpression of Four OsGRFs Enhances Rice Blast Resistance
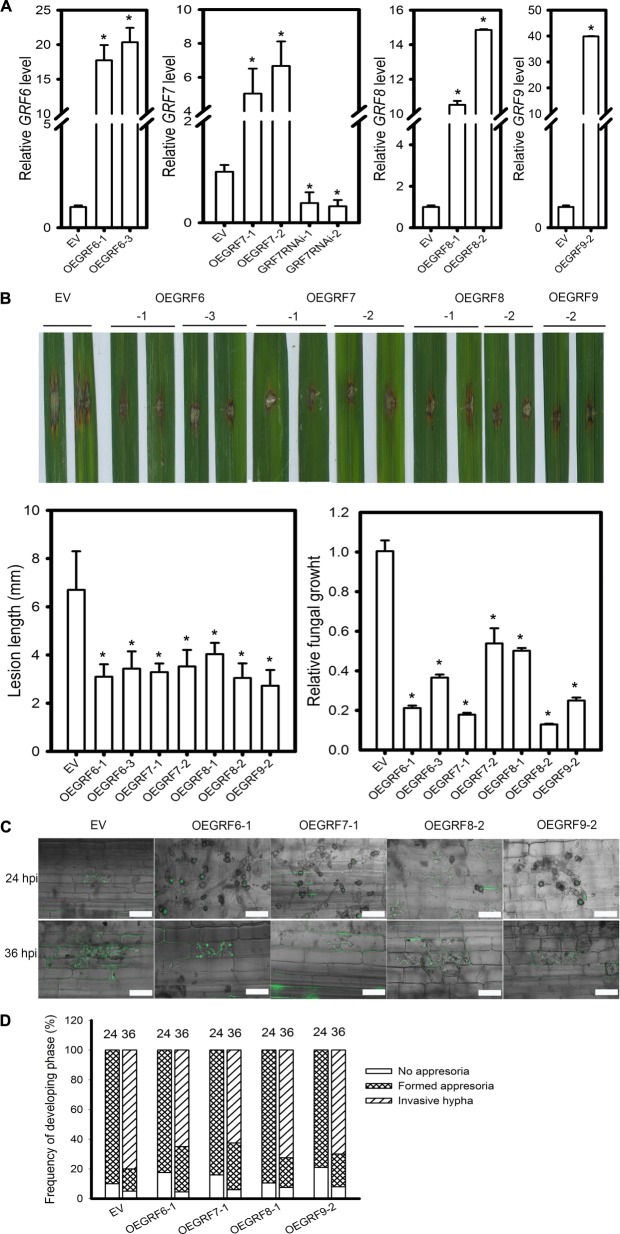
Based on the target gene expression in LTH and IRBLkm-Ts accession, we speculated that OsGRF6, OsGRF7, OsGRF8, and OsGRF9 may positively regulate rice blast resistance. Interestingly, these four genes belong to the same clade in the phylogenetic tree constructed in a previous report (Gao et al., 2016). Therefore, we made transgenic rice lines overexpressing OsGRF6, OsGRF7, OsGRF8, and OsGRF9 (OEGRF) under Yuetai-B (YB) background and performed blast disease assay. The relative expression levels of OsGRF6, OsGRF7, OsGRF8, and OsGRF9 in the respective transgenic lines were quantified (Figure 7A). All the OEGRF lines exhibited enhanced resistance to rice blast as demonstrated by the smaller lesions than those in the control plants (Figure 7B). The lesion length on the OEGRF lines was significantly reduced when compared to control plants (Figure 7B). Fungal biomass was also significantly decreased in OEGRF lines compared to control plants (Figure 7B). In addition, we examined the infection process of GZ8 on sheath cells. At 12 hpi, appressoria formation was hardly detected; whereas at 24 hpi most spores formed appressoria on control and OEGRF lines. At 36 hpi, ∼80% of spores formed invasive hyphae on control lines; in contrast, only ∼62.5 to 72.5% formed invasive hyphae on OEGRF lines (Figures 7C,D). Together, all the four OEGRF transgenic lines showed enhanced resistance to blast fungus, among which OEGRF7 was the highest resistant followed by OEGRF6, OEGRF9, and OEGRF8 (Figure 7D). To further confirm the data from OEGRF7, we examined the disease phenotypes of transgenic lines knocking-down OsGRF7 by RNA interference (GRF7RNAi). The relative OsGRF7 expression levels were quantified in transgenic lines (Figure 7A). GRF7RNAi lines exhibited enhanced susceptibility as indicated by the larger lesions and more fungal growth that the control plants (Supplementary Figure S5A). In addition, the invasive hyphae of GZ8 formed in GRF7RNAi lines were more aggressive than the control line (Supplementary Figure S5B).
FIGURE 7.
Function of four OsGRF genes in rice blast resistance. (A) RT-qPCR analyses show the relative expression of OsGRF6, OsGRF7, OsGRF8, and OsGRF9 in the indicated transgenic lines. OsUbiquitin served as the internal reference. The accumulation level was normalized to that of the control plants (EV). (B) Disease severity of the indicated lines at 5 days post inoculation of M. oryzae strain Zhong1. Punch inoculation was conducted on 4–5 week old leaves of wild type (EV) and the indicated transgenic lines. Relative fungal biomass is determined by examining the expression level of M. oryzae Pot2 gene against OsUbiquitin DNA level. (C) Representative confocal images of sheath cells from the indicated lines infected by eGFP-tagged blast isolate GZ8. Bars = 20 μm. (D) Quantification analysis on the progress of fungal infection at 24 and 36 hpi. Around 200 conidia in each line were analyzed. Similar results were obtained in at least two independent experiments. In (A,B), error bars indicate SD (n = 3). The asterisk above the bars indicate significant differences between EV and the indicated transgenic lines at a P-value < 0.01, as determined by Student’s t-test.
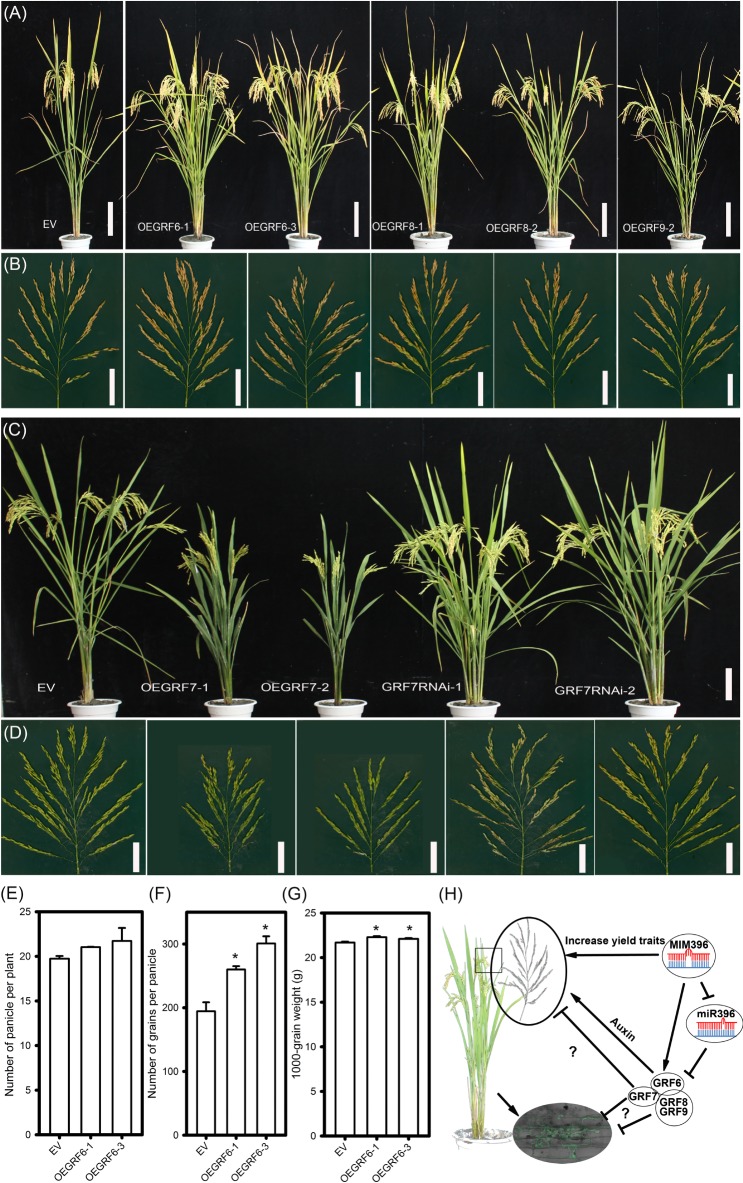
Regarding agronomic traits, among all the four OEGRF plants, OEGRF6 showed significant increase in yield-traits, such as the number of primary and secondary branches, spikelet numbers (Figures 8A,B and Supplementary Table S2) and larger grain size compared to control plants (Supplementary Table S2), consistent with the previous report (Gao et al., 2016). The number of panicle per plant, number of grains per panicle and 1000-grain weight were also increased (Figures 8E–G). In contrast, OEGRF7 showed reduced height, smaller panicles and less number of secondary branches (Figures 8C,D and Supplementary Table S2). On the contrary, GRF7RNAi lines showed phenotypes comparable to the control plants with higher number of tillers and panicles (Figures 8C,D and Supplementary Table S2). In addition, the growth in field trials was consistent at two sites (Wuhan and Chengdu, Supplementary Table S2). The performance of OEGRF6 was consistent with the earlier reports (Gao et al., 2016), whereas, OEGRF7 exhibited defects in growth. In total, the above results indicate that overexpression of OsGRF8 and OsGRF9 enhances rice blast resistance, OsGRF7 positively regulates rice blast resistance, but negatively regulates growth, and OsGRF6 positively regulates both rice blast resistance and yield traits.
FIGURE 8.
Agronomic traits of transgenic lines overexpressing different OsGRF genes and working model for miR396-OsGRF in regulation of rice immunity and yield traits. Comparison of gross plant (A), scale bar = 16 cm, and panicle morphologies (B), scale bar = 5 cm, of EV (YB), OEGRF6, OEGRF8 and OEGRF9 transgenic plants. Comparison of gross plant (C), scale bar = 16 cm, and panicle morphologies (D), scale bar = 5 cm, of EV (YB), OEGRF7, and GRF7RNAi transgenic plants. Number of panicles (E), number of grains per panicle (F), and 1000-grain weight (G) of OsGRF6 over-expression lines. The asterisk above the bars indicate significant differences between EV and the indicated transgenic lines at a P-value < 0.01, as determined by Student’s t-test. (H) Working model for miR396-OsGRFs module in balancing yield traits and immunity against the blast fungus.
Discussion
MiRNAs play important roles in regulating gene expression during plant response to pathogen infection (Seo et al., 2013; Yu et al., 2017). Recently, a large number of M. oryzae-responsive miRNAs have been identified through high throughput sequencing (Li et al., 2014; Baldrich and San Segundo, 2016; Zhang et al., 2018). As a result, miRNAs such as miR7695, miR398b, miR160a and miR166k-166h were characterized as positive regulators (Campo et al., 2013; Li et al., 2014; Salvador-Guirao et al., 2018), and miR169a, miR164a, miR319b, and miR444b.2 as negative regulators (Li Y. et al., 2017; Xiao et al., 2017; Wang et al., 2018; Zhang et al., 2018) of rice blast resistance. The miR396 family is highly conserved and contains 8 members in rice (Supplementary Figure S1). Previously, several miR396 family members were found to be responsive to the infection of M. oryzae (Campo et al., 2013; Li et al., 2014; Zhang et al., 2015). In the present study, we demonstrated that miR396 negatively regulates rice immunity against the blast fungus M. oryzae via suppression of multiple OsGRFs. Overexpression of different miR396 isoforms significantly suppressed their target genes GRFs (Figure 2B) and led to enhanced susceptibility to M. oryzae, which was attributable to reduced defense responses such as less H2O2 production (Figure 2). In contrast, blocking miR396 via target mimicry significantly up-regulated OsGRFs and led to enhanced resistance against M. oryzae (Figure 5). The expression of multiple tested OsGRF target genes was suppressed at transcriptional level by overexpressing miR396 isoforms (Figures 2B, 3, 4), but upregulated in MIM396 lines (Figure 5B), suggesting that the role of miR396 is through manipulating the expression of OsGRFs. Consistently, the transgenic plants overexpressing OsGRF6, OsGRF7, OsGRF8, and OsGRF9 displayed enhanced resistance against M. oryzae (Figure 7). The expression patterns of miR396 also support its negative roles in rice immunity against M. oryzae. The accumulation of miR396 isoforms was increased consistently at all the time points in the susceptible accession LTH; however, their expression levels were fluctuated in IRBLkm-Ts (Figure 1). In a previous report (Campo et al., 2013), miR396c expression was decreased, whereas, its target genes LOC_Os06g10310 (OsGRF2) and LOC_Os03g51970 (OsGRF6) were increased upon treatment with elicitors of M. oryzae. Similarly, miR396e expression was also down regulated upon infection of M. oryzae (Zhang et al., 2015). Thus, the above data demonstrate that miR396 negatively regulates rice blast resistance.
To date, a number of transcription factors, such as WRKY, NAM/ATAF/CUC (NAC), ethylene responsive factor/APETALA2 (ERF/AP2), basic helix-loop helix (bHLH), basic-domain leucine-zipper (bZIP) and MADS box, have been identified to be involved in regulating immunity in crops (Ning et al., 2017). MiR396 targets GRFs, which encode transcription factors that involve in plant growth and development (Omidbakhshfard et al., 2015). Accumulating evidence showed that GRFs may play roles in defense responses (Liu J.Y. et al., 2014; Xu et al., 2014; Chen et al., 2015). Here, our data showed that OsGRFs were differentially responsive to M. oryzae infection in LTH and IRBLKm-Ts (Figure 6). These genes were divided into three clades based on the similarity in amino acid sequences (Gao et al., 2016). Namely OsGRF1, OsGRF2, OsGRF3, OsGRF4, and OsGRF5 belong to clade 1; OsGRF6, OsGRF7, OsGRF8, and OsGRF9 belong to clade 2; and OsGRF10, OsGRF11, and OsGRF12 belong to the clade 3 (Gao et al., 2016). Interestingly, the expression level of genes in the clade 1 (OsGRF2, OsGRF3, OsGRF4, and OsGRF5) and clade 3 (OsGRF11) were slightly increased (Figures 6A,B), whereas, increased remarkably for the genes in the clade 2 (Figures 6C,D) upon M. oryzae infection, indicating that genes of clade 2 (OsGRF6, OsGRF7, OsGRF8, and OsGRF9) are highly responsive in comparison to other clades upon M. oryzae infection. In addition, OsGRF6, OsGRF7, OsGRF8, and OsGRF9 might function as positive regulators because their expression levels were significantly up-regulated in the resistant accession IRBLkm-Ts upon M. oryzae infection (Figure 6D). Consistently, all the transgenic OEGRF lines from clade 2 exhibited enhanced resistance to M. oryzae (Figure 7). Among them, OEGRF7 showed the highest resistance (Figure 7D), which was further confirmed by enhanced susceptibility to M. oryzae in GRF7RNAi lines (Supplementary Figure S5). Altogether, the above data describes a novel function of GRFs in rice immunity against rice blast fungus.
In rice, different isoforms of miR396 preferentially regulates different GRFs involving multiple functions. For example, miR396c decreases salt and alkali stress tolerance by regulating OsGRF10 and OsGRF3 (Gao et al., 2010). miR396d affects spikelet development by regulating the expression of OsGRF6 and OsGRF10 (Liu H.H. et al., 2014). MiR396b regulates OsGRF6 for increased grain yield by modulating development of auxiliary branches and spikelets (Gao et al., 2016). Moreover, miR396d (Che et al., 2016), miR396g/h (Duan et al., 2016), miR396c (Hu et al., 2015; Li et al., 2016), regulates OsGRF4 for controlling grain size and yield, respectively. Here, miR396a, miR396c, miR396d, and miR396h affect yield traits such as seed length, width and 1000-grain weight (Supplementary Table S1) by suppressing six to eight OsGRFs (Figure 2B), respectively, consistent with the previous report (Li et al., 2016). Moreover, MIM396 and OEGRF6 lines showed significant improvement in yield traits such as secondary branches, spikelet numbers and grain size (Figures 5, 8 and Supplementary Table S2), confirming the involvement of miR396-OsGRF6 module in controlling the development of secondary branches in rice inflorescences as suggested in a previous study (Gao et al., 2016). Likewise, miR156 and miR397 have been found to control rice yield. MiR156 regulates rice yield by fine tuning the expression of SPL genes (Wang and Zhang, 2017). MiR397 regulates rice yield by increasing spikelet number and grain size by targeting OsLAC (Zhang et al., 2013; Zheng and Qu, 2015).
High yield and resistance to pathogens are the major goals in plant breeding. But, they usually act antagonistically and in many cases, the genes that are well known to play defense roles often comes with compromise in plant growth (Ning et al., 2017). For example, mutation in SPL28 showed enhanced resistance to M. oryzae, but led to reduction in yield (Qiao et al., 2010). However, several recent studies have reported that better disease resistance and improved yield could be simultaneously achieved. For example, the paired NBS-LRR receptors, PigmR and PigmS, confer high rice blast disease resistance without yield penalty via an epigenetic regulatory mechanism (Deng et al., 2017). Using pathogen-inducible upstream open reading frame (uORF)-mediated translational control over AtNPR1, Xu et al. (2017) obtained engineered rice conferring broad-spectrum disease resistance without fitness costs. Notably, the rice transcription factor Ideal Plant Architecture 1 (IPA1) could improve both grain yield and blast disease resistance through switching DNA binding specificity controlled by phosphorylation (Wang et al., 2018). Furthermore, miRNAs could be ideal candidate to promote both immunity and yield, by fine tuning the expression of their target genes. For example, overexpressing miR444b.1 negatively regulates rice blast resistance with reduced tiller numbers (Xiao et al., 2017), whereas, overexpression of its target gene OsMADS57 resulted in increased number of tillers (Guo et al., 2013). Similarly, miR164a negatively regulates M. oryzae resistance by suppressing OsNAC60 (Wang et al., 2018), whereas overexpression of another miR164 target, OsNAC2, leads to improvement of plant architecture and grain yield in rice (Jiang et al., 2018). In these reports, immunity and yield traits were regulated by different miRNA isoforms or different target genes. Here, we demonstrated a new role for four OsGRFs that positively regulated resistance to M. oryzae, in addition to their balanced regulation of yield traits (Figures 7, 8). Collectively, we showed that miR396-OsGRFs module regulates the trade-offs between plant immunity and yield (Figure 8H). On one hand, miR396 negatively regulates blast disease resistance through suppression of OsGRF6, OsGRF7, OsGRF8, and OsGRF9, which in turn, positively regulate blast disease resistance and coordinate development via a yet identify mechanism. Thus, our data provide a potential regulatory module in improvement of blast disease-resistance and yield in rice breeding programs. Since miR396-GRFs module is highly conserved in plants (Liu et al., 2009, 2017; Debernardi et al., 2012; Bazin et al., 2013; Gao et al., 2016), its roles in balancing yield and immunity might be applied in breeding programs for crop improvement.
Author Contributions
VC, HW, FG, X-LC, Y-PC, G-BL, YZ, X-MY, L-LZ, Z-XZ, J-HZ, Y-GW, and SL performed the experiments. JF, J-QZ, and YL assisted in experiments. VC and W-MW wrote the manuscript. W-MW and S-QL coordinated the overall study and edited the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This work was supported by the National Natural Science Foundation of China (Grants 31430072 and 31672090 to W-MW, 31471761 to YL, and 31570004 to SL) and National Transgenic Research Program of China (2016ZX08001004-001-002 to S-QL).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.01999/full#supplementary-material
Basic information of miR396 family members. (A) Chromosomal location of the eight miR396 family members in rice. The centromere in each chromosome is indicated by a circle. (B) Phylogenetic tree derived from the nucleotide sequence comparison. The tree was generated using MEGA 5 by the CLUSTAL W method. The miRBase accession number of the sequences used in the comparison is shown near the miRNA ID. The phylogenetic tree was generated as a consensus of 1,000 bootstrap replicates by the neighbor-joining method. (C) Comparison of the mature sequences of miR396 isoforms from miRBase.
Alignment of miR396d with target sequences.
Overexpression of miR396 target mimicry results in enhanced resistance to M. oryzae (NC10). Punch inoculation of 4–5 week old leaves from YB (WT), MIM396-1 and MIM396-2 show disease severity of M. oryzae (1 × 105 spore/ml conc., NC10) at 5 days post inoculation. Error bars indicate SD from three biological replicates. The letters above the bars indicate significant differences between (EV) and the indicated lines at a P-value < 0.01, as determined by a one-way ANOVA followed by post hoc Tukey HSD analysis.
Overexpression of STTM396 results in enhanced resistance to M. oryzae. Punch inoculation of 4–5 week old leaves from NPB (EV), STTM396-1 and STTM396-2 show disease severity of M. Oryzae (1 × 105 spore/ml conc., Zhong1) at 5 days post inoculation. Error bars indicate SD from three biological replicates. The letters above the bars indicate significant differences between EV and the indicated lines at a P-value < 0.01, as determined by a one-way ANOVA followed by post hoc Tukey HSD analysis.
Silencing of OsGRF7 resulted in enhanced susceptibility to rice blast fungus. (A) Punch inoculation of 4–5 week old leaves from EV (YB) and GRF7RNAi rice plants show disease severity of M. oryzae (Zhong1, 1 × 105 spore/ml conc.) at 5 days post inoculation. Relative fungal biomass is determined by examining the expression level of M. oryzae Pot2 gene against OsUbiqutin DNA level. (B) Representative confocal images of EV (YB) and GRF7RNAi-1 sheath cells infected by eGFP-tagged blast isolate GZ8, Bars = 20 μm.
Agronomic performances of miR396 over expression lines.
Agronomic performances of MIM396 and OEGRF transgenic lines.
Primers used in this study.
References
- Baldrich P., Campo S., Wu M. T., Liu T. T., Hsing Y. L., San Segundo B. (2015). MicroRNA-mediated regulation of gene expression in the response of rice plants to fungal elicitors. RNA Biol. 12 847–863. 10.1080/15476286.2015.1050577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldrich P., San Segundo B. (2016). MicroRNAs in rice innate immunity. Rice 9:6. 10.1186/S12284-016-0078-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazin J., Khan G. A., Combier J. P., Bustos-Sanmamed P., Debernardi J. M., Rodriguez R., et al. (2013). miR396 affects mycorrhization and root meristem activity in the legume Medicago truncatula. Plant J. 74 920–934. 10.1111/tpj.12178 [DOI] [PubMed] [Google Scholar]
- Campo S., Peris-Peris C., Sire C., Moreno A. B., Donaire L., Zytnicki M., et al. (2013). Identification of a novel microRNA (miRNA) from rice that targets an alternatively spliced transcript of the Nramp6 (natural resistance-associated macrophage protein 6) gene involved in pathogen resistance. New Phytol. 199 212–227. 10.1111/nph.12292 [DOI] [PubMed] [Google Scholar]
- Che R. H., Tong H. N., Shi B. H., Liu Y. Q., Fang S. R., Liu D. P., et al. (2016). Control of grain size and rice yield by GL2-mediated brassinosteroid responses. Nat. Plants 2:15195. 10.1038/Nplants2016.2 [DOI] [PubMed] [Google Scholar]
- Chen H. M., Zou Y., Shang Y. L., Lin H. Q., Wang Y. J., Cai R., et al. (2008). Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol. 146 368–376. 10.1104/pp.107.111740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Luan Y. S., Zhai J. M. (2015). Sp-miR396a-5p acts as a stress-responsive genes regulator by conferring tolerance to abiotic stresses and susceptibility to Phytophthora nicotianae infection in transgenic tobacco. Plant Cell Rep. 34 2013–2025. 10.1007/s00299-015-1847-0 [DOI] [PubMed] [Google Scholar]
- Chen X. W., Ronald P. C. (2011). Innate immunity in rice. Trends Plant Sci. 16 451–459. 10.1016/j.tplants.2011.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debernardi J. M., Rodriguez R. E., Mecchia M. A., Palatnik J. F. (2012). Functional specialization of the plant miR396 regulatory network through distinct microRNA-target interactions. PLoS Genet. 8:e1002419. 10.1371/journal.pgen.1002419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y., Wang J., Tung J., Liu D., Zhou Y., He S., et al. (2018). A role for small RNA in regulating innate immunity during plant growth. PLoS Pathog. 14:e1006756. 10.1371/journal.ppat.1006756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y. W., Zhai K. R., Xie Z., Yang D. Y., Zhu X. D., Liu J. Z., et al. (2017). Epigenetic regulation of antagonistic receptors confers rice blast resistance with yield balance. Science 355 962–965. 10.1126/science.aai8898 [DOI] [PubMed] [Google Scholar]
- Duan P. G., Ni S., Wang J. M., Zhang B. L., Xu R., Wang Y. X., et al. (2016). Regulation of OsGRF4 by OsmiR396 controls grain size and yield in rice. Nat. Plants 2:15203. 10.1038/Nplants.2015.203 [DOI] [PubMed] [Google Scholar]
- Fahlgren N., Howell M. D., Kasschau K. D., Chapman E. J., Sullivan C. M., Cumbie J. S., et al. (2007). High-throughput sequencing of arabidopsis microRNAs: evidence for frequent birth and death of MIRNA genes. PLoS One 2:e219. 10.1371/journal.pone.0000219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco-Zorrilla J. M., Valli A., Todesco M., Mateos I., Puga M. I., Rubio-Somoza I., et al. (2007). Target mimicry provides a new mechanism for regulation of microRNA activity. Nat. Genet. 39 1033–1037. 10.1038/ng2079 [DOI] [PubMed] [Google Scholar]
- Gao F., Wang K., Liu Y., Chen Y. P., Chen P. A., Shi Z. Y., et al. (2016). Blocking miR396 increases rice yield by shaping inflorescence architecture. Nat. Plants 2:15196. 10.1038/Nplants.2015.196 [DOI] [PubMed] [Google Scholar]
- Gao P., Bai X., Yang L., Lv D. K., Li Y., Cai H., et al. (2010). Over-expression of osa-MIR396c decreases salt and alkali stress tolerance. Planta 231 991–1001. 10.1007/s00425-010-1104-2 [DOI] [PubMed] [Google Scholar]
- Guo S. Y., Xu Y. Y., Liu H. H., Mao Z. W., Zhang C., Ma Y., et al. (2013). The interaction between OsMADS57 and OsTB1 modulates rice tillering via DWARF14. Nat. Commun. 4:1566. 10.1038/ncomms2542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J., Wang Y. X., Fang Y. X., Zeng L. J., Xu J., Yu H. P., et al. (2015). A rare allele of GS2 enhances grain size and grain yield in rice. Mol. Plant 8 1455–1465. 10.1016/j.molp.2015.07.002 [DOI] [PubMed] [Google Scholar]
- Huang Y. Y., Shi Y., Lei Y., Li Y., Fan J., Xu Y. J., et al. (2014). Functional identification of multiple nucleocytoplasmic trafficking signals in the broad-spectrum resistance protein RPW8.2. Planta 239 455–468. 10.1007/s00425-013-1994-x [DOI] [PubMed] [Google Scholar]
- Jagadeeswaran G., Saini A., Sunkar R. (2009). Biotic and abiotic stress down-regulate miR398 expression in arabidopsis. Planta 229 1009–1014. 10.1007/s00425-009-0889-3 [DOI] [PubMed] [Google Scholar]
- Jiang D. G., Chen W. T., Dong J. F., Li J., Yang F., Wu Z. C., et al. (2018). Over expression of miR164b-resistant OsNAC2 improves plant architecture and grain yield in rice. J. Exp. Bot. 69 1533–1543. 10.1093/jxb/ery017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kankanala P., Czymmek K., Valent B. (2007). Roles for rice membrane dynamics and plasmodesmata during biotrophic invasion by the blast fungus. Plant Cell 19 706–724. 10.1105/tpc.106.046300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Pignatta D., Bendix C., Brunkard J. O., Cohn M. M., Tung J., et al. (2012). MicroRNA regulation of plant innate immune receptors. Proc. Natl. Acad. Sci. U.S.A. 109 1790–1795. 10.1073/pnas.1118282109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S. C., Gao F. Y., Xie K. L., Zeng X. H., Cao Y., Zeng J., et al. (2016). The OsmiR396c-OsGRF4-OsGIF1 regulatory module determines grain size and yield in rice. Plant Biotechnol. J. 14 2134–2146. 10.1111/pbi.12569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W. T., Zhu Z. W., Chern M. S., Yin J. J., Yang C., Ran L., et al. (2017). A natural allele of a transcription factor in rice confers broad-spectrum blast resistance. Cell 170 114.e15–126.e126. 10.1016/j.cell.2017.06.008 [DOI] [PubMed] [Google Scholar]
- Li Y., Zhao S. L., Li J. L., Hu X. H., Wang H., Cao X. L., et al. (2017). Osa-miR169 negatively regulates rice immunity against the blast fungus Magnaporthe oryzae. Front. Plant Sci. 8:2. 10.3389/Fpls.2017.00002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Lu Y. G., Shi Y., Wu L., Xu Y. J., Huang F., et al. (2014). Multiple rice microRNAs are involved in immunity against the blast fungus Magnaporthe oryzae. Plant Physiol. 164 1077–1092. 10.1104/pp.113.230052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Zhang Q. Q., Zhang J. G., Wu L., Qi Y. J., Zhou J. M. (2010). Identification of microRNAs involved in pathogen-associated molecular pattern-triggered plant innate immunity. Plant Physiol. 152 2222–2231. 10.1104/pp.109.151803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D. M., Song Y., Chen Z. X., Yu D. Q. (2009). Ectopic expression of miR396 suppresses GRF target gene expression and alters leaf growth in arabidopsis. Physiol. Plant. 136 223–236. 10.1111/j.1399-3054.2009.01229.x [DOI] [PubMed] [Google Scholar]
- Liu H. H., Guo S. Y., Xu Y. Y., Li C. H., Zhang Z. Y., Zhang D. J., et al. (2014). OsmiR396d-regulated OsGRFs function in floral organogenesis in rice through binding to their targets OsJMJ706 and OsCR4. Plant Physiol. 165 160–174. 10.1104/pp.114.235564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J. Y., Rice J. H., Chen N. N., Baum T. J., Hewezi T. (2014). Synchronization of developmental processes and defense signaling by growth regulating transcription factors. PLoS One 9:e98477. 10.1371/journal.pone.0098477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W., Zhou Y., Li X., Wang X., Dong Y., Wang N., et al. (2017). Tissue-specific regulation of gma-miR396 family on coordinating development and low water availability responses. Front. Plant Sci. 8:1112. 10.3389/fpls.2017.01112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C., Lu Y., Bai S. L., Zhang W. N., Duan X. W., Meng D., et al. (2014). Cloning and characterization of miRNAs and their targets, including a novel miRNA-targeted NBS-LRR protein class gene in apple (golden delicious). Mol. Plant 7 218–230. 10.1093/mp/sst101 [DOI] [PubMed] [Google Scholar]
- Navarro L., Dunoyer P., Jay F., Arnold B., Dharmasiri N., Estelle M., et al. (2016). A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science 352 1286–1286. 10.1126/science.1126088 [DOI] [PubMed] [Google Scholar]
- Ning Y. S., Liu W. D., Wang G. L. (2017). Balancing immunity and yield in crop plants. Trends Plant Sci. 22 1069–1079. 10.1016/j.tplants.2017.09.010 [DOI] [PubMed] [Google Scholar]
- Omidbakhshfard M. A., Proost S., Fujikura U., Mueller-Roeber B. (2015). Growth-regulating factors (GRFs): a small transcription factor family with important functions in plant biology. Mol. Plant 8 998–1010. 10.1016/j.molp.2015.01.013 [DOI] [PubMed] [Google Scholar]
- Park C. H., Chen S. B., Shirsekar G., Zhou B., Khang C. H., Songkumarn P., et al. (2012). The Magnaporthe oryzae effector AvrPiz-t targets the Ring E3 ubiquitin ligase APIP6 to suppress pathogen-associated molecular pattern-triggered immunity in rice. Plant Cell 24 4748–4762. 10.1105/tpc.112.105429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao Y., Jiang W., Lee J., Park B., Choi M. S., Piao R., et al. (2010). SPL28 encodes a clathrin-associated adaptor protein complex 1, medium subunit mu 1 (AP1M1) and is responsible for spotted leaf and early senescence in rice (Oryza sativa). New Phytol. 185 258–274. 10.1111/j.1469-8137.2009.03047.x [DOI] [PubMed] [Google Scholar]
- Ramirez-Prado J. S., Abulfaraj A. A., Rayapuram N., Benhamed M., Hirt H. (2018). Plant immunity: from signaling to epigenetic control of defense. Trends Plant Sci. 23 833–844. 10.1016/j.tplants.2018.06.004 [DOI] [PubMed] [Google Scholar]
- Salvador-Guirao R., Hsing Y. I., San Segundo B. (2018). The polycistronic miR166k-166h positively regulates rice immunity via post-transcriptional control of EIN2. Front. Plant Sci. 9:337. 10.3389/fpls.2018.00337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo J. K., Wu J. G., Lii Y. F., Li Y., Jin H. L. (2013). Contribution of small RNA pathway components in plant immunity. Mol. Plant Microbe Interact. 26 617–625. 10.1094/Mpmi-10-12-0255-Ia [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivaprasad P. V., Chen H. M., Patel K., Bond D. M., Santos B. A., Baulcombe D. C. (2012). A microRNA super family regulates nucleotide binding site-leucine-rich repeats and other mRNAs. Plant Cell 24 859–874. 10.1105/tpc.111.095380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto-Suarez M., Baldrich P., Weigel D., Rubio-Somoza I., San Segundo B. (2017). The arabidopsis miR396 mediates pathogen-associated molecular pattern-triggered immune responses against fungal pathogens. Sci. Rep. 7:44898. 10.1038/Srep44898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y. Y., Liu H. H., Guo S. Y., Wang B., Li Z. T., Chong K., et al. (2018). OsmiR396d affects gibberellin and brassinosteroid signaling to regulate plant architecture in rice. Plant Physiol. 176 946–959. 10.1104/pp.17.00964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner M., Adhikari S., Subramanian S. (2013). Optimizing stem-loop qPCR assays through multiplexed cDNA synthesis of U6 and miRNAs. Plant Signal Behav. 8:e24918. 10.4161/psb.24918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varkonyi-Gasic E., Wu R. M., Wood M., Walton E. F., Hellens R. P. (2007). Protocol: a highly sensitive RT-PCR method for detection and quantification of microRNAs. Plant Methods 3:12. 10.1186/1746-4811-3-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Zhou L., Shi H., Chern M., Yu H., Yi H., et al. (2018). A single transcription factor promotes both yield and immunity in rice. Science 361 1026–1028. 10.1126/science.aat7675 [DOI] [PubMed] [Google Scholar]
- Wang L., Zhang Q. F. (2017). Boosting rice yield by fine-tuning SPL gene expression. Trends Plant Sci. 22 643–646. 10.1016/j.tplants.2017.06.004 [DOI] [PubMed] [Google Scholar]
- Wu F. L., Guo Q. L., Zhang W., Jin W. B. (2015). Identification and analysis of powdery mildew-responsive mirnas in wheat. J. Phytopathol. 163 264–270. 10.1111/jph.12315 [DOI] [Google Scholar]
- Xiao S., Brown S., Patrick E., Brearley C., Turner J. G. (2003). Enhanced transcription of the Arabidopsis disease resistance genes RPW8.1 and RPW8.2 via a salicylic acid-dependent amplification circuit is required for hypersensitive cell death. Plant Cell 15 33–45. 10.1105/tpc.006940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Z. Y., Wang Q. X., Wang H., Li J. L., Zhao S. L., Fan J., et al. (2017). MiR444b.2 regulates resistance to Magnaporthe oryzae and tillering in rice. Acta Phytopathol. Sin. 47 511–522. 10.13926/j.cnki.apps.000019 [DOI] [Google Scholar]
- Xin M. M., Wang Y., Yao Y. Y., Xie C. J., Peng H. R., Ni Z. F., et al. (2010). Diverse set of microRNAs are responsive to powdery mildew infection and heat stress in wheat (Triticum aestivum L.). BMC Plant Biol. 10:123. 10.1186/1471-2229-10-123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D. L., Mou G. P., Wang K., Zhou G. H. (2014). MicroRNAs responding to southern rice black-streaked dwarf virus infection and their target genes associated with symptom development in rice. Virus Res. 190 60–68. 10.1016/j.virusres.2014.07.007 [DOI] [PubMed] [Google Scholar]
- Xu G. Y., Uan M. Y., Ai C. R., Liu L. J., Zhuang E., Karapetyan S., et al. (2017). uORF-mediated translation allows engineered plant disease resistance without fitness costs. Nature 545 491–494. 10.1038/nature22372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y., Jia T. R., Chen X. M. (2017). The ‘how’ and ‘where’ of plant microRNAs. New Phytol. 216 1002–1017. 10.1111/nph.14834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D. D., Liu M. X., Tang M. Z., Dong B., Wu D. X., Zhang Z. G., et al. (2015). Repression of microRNA biogenesis by silencing of OsDCL1 activates the basal resistance to Magnaporthe oryzae in rice. Plant Sci. 237 24–32. 10.1016/j.plantsci.2015.05.002 [DOI] [PubMed] [Google Scholar]
- Zhang X., Bao Y., Shan D., Wang Z., Song X., Wang J., et al. (2018). Magnaporthe oryzae defeats rice defense by inducing miR319b and suppressing jasmonic acid signaling. Plant Physiol. 177 352–368. 10.1104/pp.17.01665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. C., Yu Y., Wang C. Y., Li Z. Y., Liu Q., Xu J., et al. (2013). Overexpression of microRNA OsmiR397 improves rice yield by increasing grain size and promoting panicle branching. Nat. Biotechnol. 31 848–852. 10.1038/nbt.2646 [DOI] [PubMed] [Google Scholar]
- Zheng L. L., Qu L. H. (2015). Application of microRNA gene resources in the improvement of agronomic traits in rice. Plant Biotechnol. J. 13 329–336. 10.1111/pbi.12321 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Basic information of miR396 family members. (A) Chromosomal location of the eight miR396 family members in rice. The centromere in each chromosome is indicated by a circle. (B) Phylogenetic tree derived from the nucleotide sequence comparison. The tree was generated using MEGA 5 by the CLUSTAL W method. The miRBase accession number of the sequences used in the comparison is shown near the miRNA ID. The phylogenetic tree was generated as a consensus of 1,000 bootstrap replicates by the neighbor-joining method. (C) Comparison of the mature sequences of miR396 isoforms from miRBase.
Alignment of miR396d with target sequences.
Overexpression of miR396 target mimicry results in enhanced resistance to M. oryzae (NC10). Punch inoculation of 4–5 week old leaves from YB (WT), MIM396-1 and MIM396-2 show disease severity of M. oryzae (1 × 105 spore/ml conc., NC10) at 5 days post inoculation. Error bars indicate SD from three biological replicates. The letters above the bars indicate significant differences between (EV) and the indicated lines at a P-value < 0.01, as determined by a one-way ANOVA followed by post hoc Tukey HSD analysis.
Overexpression of STTM396 results in enhanced resistance to M. oryzae. Punch inoculation of 4–5 week old leaves from NPB (EV), STTM396-1 and STTM396-2 show disease severity of M. Oryzae (1 × 105 spore/ml conc., Zhong1) at 5 days post inoculation. Error bars indicate SD from three biological replicates. The letters above the bars indicate significant differences between EV and the indicated lines at a P-value < 0.01, as determined by a one-way ANOVA followed by post hoc Tukey HSD analysis.
Silencing of OsGRF7 resulted in enhanced susceptibility to rice blast fungus. (A) Punch inoculation of 4–5 week old leaves from EV (YB) and GRF7RNAi rice plants show disease severity of M. oryzae (Zhong1, 1 × 105 spore/ml conc.) at 5 days post inoculation. Relative fungal biomass is determined by examining the expression level of M. oryzae Pot2 gene against OsUbiqutin DNA level. (B) Representative confocal images of EV (YB) and GRF7RNAi-1 sheath cells infected by eGFP-tagged blast isolate GZ8, Bars = 20 μm.
Agronomic performances of miR396 over expression lines.
Agronomic performances of MIM396 and OEGRF transgenic lines.
Primers used in this study.