Human herpesvirus 6B (HHV-6B) is a DNA virus that infects most children within the first few years of life. After primary infection, HHV-6B persists as a chronic, latent infection in many cell types. Additionally, HHV-6B can integrate into germ line chromosomes, resulting in individuals with viral DNA in every nucleated cell. Given that PCR to detect viral DNA is the mainstay for diagnosing HHV-6B infection, the characteristics of HHV-6B infection complicate efforts to distinguish between latent and active viral infection, particularly in immunocompromised patients who have frequent HHV-6B reactivation. In this study, we used RNA sequencing to characterize the HHV-6B gene expression profile in multiple sample types, and our findings identified evidence-based targets for diagnostic tests that distinguish between latent and active viral infection.
KEYWORDS: herpes, HHV-6, transcriptome, RNA sequencing, RNA-seq, transplant, diagnose, bone marrow transplantation, diagnostics, gene sequencing, human herpesviruses, transcription, transplant infectious diseases
ABSTRACT
Human herpesvirus 6B (HHV-6B) DNA is frequently detected in human samples. Diagnostic assays distinguishing HHV-6B reactivation from latency are limited. This has impaired strategies to diagnose and treat HHV-6B-associated diseases. We used RNA sequencing to characterize and compare the HHV-6B transcriptome in multiple sample types, including (i) whole blood from hematopoietic cell transplant (HCT) recipients with and without HHV-6B plasma viremia, (ii) tumor tissue samples from subjects with large B cell lymphoma infected with HHV-6B, (iii) lymphoblastoid cell lines (LCLs) from subjects with inherited chromosomally integrated HHV-6B or latent infection with HHV-6B, and (iv) HHV-6B Z29 infected SupT1 CD4+ T cells. We demonstrated substantial overlap in the HHV-6B transcriptome observed in in vivo and in vitro samples, although there was variability in the breadth and quantity of gene expression across samples. The HHV-6B viral polymerase gene U38 was the only HHV-6B transcript detected in all next-generation RNA sequencing (RNA-seq) data sets and was one of the most highly expressed genes. We developed a novel reverse transcription-PCR assay targeting HHV-6B U38, which identified U38 mRNA in all tested whole-blood samples from patients with concurrent HHV-6B viremia. No HHV-6B U38 transcripts were detected by RNA-seq or reverse transcription–real-time quantitative PCR (RT-qPCR) in whole-blood samples from subjects without HHV-6B plasma detection or from latently infected LCLs. A RT-qPCR assay for HHV-6B U38 may be useful to identify lytic HHV-6B infection in nonplasma samples and samples from individuals with inherited chromosomally integrated HHV-6B. This study also demonstrates the feasibility of transcriptomic analyses for HCT recipients.
IMPORTANCE Human herpesvirus 6B (HHV-6B) is a DNA virus that infects most children within the first few years of life. After primary infection, HHV-6B persists as a chronic, latent infection in many cell types. Additionally, HHV-6B can integrate into germ line chromosomes, resulting in individuals with viral DNA in every nucleated cell. Given that PCR to detect viral DNA is the mainstay for diagnosing HHV-6B infection, the characteristics of HHV-6B infection complicate efforts to distinguish between latent and active viral infection, particularly in immunocompromised patients who have frequent HHV-6B reactivation. In this study, we used RNA sequencing to characterize the HHV-6B gene expression profile in multiple sample types, and our findings identified evidence-based targets for diagnostic tests that distinguish between latent and active viral infection.
INTRODUCTION
Human herpesvirus 6B (HHV-6B) is a double-stranded DNA virus that infects the majority of people during infancy and establishes latency in a diverse array of cell types (1). Immunosuppression due to allogeneic hematopoietic cell transplantation (HCT) results in frequent viral reactivation with detection of HHV-6B DNA viremia in approximately 40% to 60% of patients. HHV-6B reactivation is the most frequent cause of encephalitis in HCT recipients, affecting 1% to 10% of patients (2), and HHV-6B is associated with a variety of disease processes in both immunocompromised and immunocompetent individuals (1). Quantitative PCR (qPCR) assays to detect HHV-6B genomic DNA in biologic samples are limited by the detection of latent virus and low specificity for end-organ disease (3–7). This is especially problematic in patients with inherited chromosomally integrated HHV-6B (iciHHV-6B), who have a high burden of cell-associated HHV-6 DNA due to integration of the viral genome in every nucleated cell (8, 9). These limitations in identification of HHV-6B reactivation and associated pathogenicity confound research and treatment efforts for HHV-6B (10).
Molecular detection of HHV-6B gene transcripts using reverse transcription–real-time quantitative PCR (RT-qPCR) has potential to improve the specificity of diagnostic studies for HHV-6B reactivation in nonplasma samples and in individuals with iciHHV-6B. This method of amplifying mRNA from infected cells may provide a better approach to distinguish active from latent infections. A few studies have reported the development and application of assays to detect HHV-6 mRNA transcripts, including open reading frames (ORFs) U12, U16/17, U60/66, U79/U80, U89/90, and U100 (11–17). However, these studies have been limited by reliance on in vitro data for transcript selection, use of selected mRNA targets based on spliced gene products, and suboptimal RNA preservation due to sample collection methods. High-throughput qualitative profiling of transcript changes using microarrays has been used to identify HHV-6B gene expression with in vitro cell lines in one study (18), but these methods may also produce biased results through the use of predefined probe sets. To our knowledge, only one study performed HHV-6B transcriptome analysis with unbiased next-generation RNA sequencing (RNA-seq) in tissue samples from two patients with diffuse large B-cell lymphoma (DLBCL) (19).
We sought to characterize in vivo HHV-6B gene expression using high-throughput RNA-seq on poly(A)-selected RNA isolated from whole-blood samples from HCT recipients with HHV-6B reactivation. To facilitate identification of highly expressed mRNA transcripts in biologic specimens to guide development of diagnostic assays, we compared HHV-6B gene expression patterns in whole blood to data from a study using tumor tissue samples and to cell cultures with lytic HHV-6B, latent HHV-6B, and iciHHV-6B. We also assessed whether we could identify distinct host gene transcription and cytokine profiles among HCT recipients with and without HHV-6B DNA detection in plasma samples.
(This article was submitted to an online preprint archive [20].)
RESULTS
Patients.
We identified seven subjects (cases 1 to 7) with whole-blood PAXgene tubes obtained post-HCT who had concurrent HHV-6B DNA detection in plasma by qPCR with a median of 4 days (range, 0 to 11) between plasma detection and PAXgene tube collection. HHV-6B was detected in plasma samples prior to, on the day of, and after the day PAXgene tube collection in 2, 1, and 3 cases, respectively; HHV-6B was detected prior to and after the day of PAXgene tube collection in 1 case (Table 1). Matched controls who had a PAXgene tube collected and did not have HHV-6B DNA plasma detection within 7 days of the PAXgene tube collection were identified for cases 1 to 5. Demographics and clinical characteristics of the cohort are presented in Table 2. None of the subjects developed HHV-6-associated end-organ disease. In addition, we obtained RNA-seq data sets from two patients with DLBCL who had HHV-6B gene expression demonstrated in tumor tissue samples (19).
TABLE 1.
Post-hematopoietic cell transplant day of HHV-6B DNA plasma detection relative to day of whole-blood PAXgene tube collection
| Case no. |
PAXgene daya |
Closest plasma HHV-6B detection daya |
Closest HHV-6B plasma viral loadb |
|---|---|---|---|
| 1 | 26 | 20 (−6) | 6,900 |
| 2 | 22 | 22 (0) | 2,000 |
| 3 | 17 | 21 (+4) | 1,567 |
| 4 | 31 | 27 (−4) | 518 |
| 5 | 7 | 18 (+11) | 2,012 |
| 6 | 13 | 17 (+4) | 4,827 |
| 7 | 26 | 25c (−1) | 4,057 |
Day post-HCT. The number in parentheses is the number of days the plasma sample with HHV-6 detection was obtained before (−) or after (+) the PAXgene tube.
Data are presented as copies per milliliter.
This subject also had HHV-6B DNA detected on day 32 (+6) with a viral load of 364 copies/ml.
TABLE 2.
Demographic and clinical characteristics of post-hematopoietic cell transplant cases (with HHV-6B plasma detection) and controls (without HHV-6B plasma detection) with whole blood collected in PAXgene tubesa
| Subject | Age (yrs) | Sex | Race | Disease | Donor | HLA matchb | Engraftment dayd ,e | GVHD dayd ,f |
|---|---|---|---|---|---|---|---|---|
| Case 1 | 46 | Male | Caucasian | AML | Unrelated | Mismatchedc | 17 | |
| Control 1 | 56 | Male | Hispanic | AML | Unrelated | Mismatchedc | 8 | 47 |
| Case 2 | 59 | Male | Caucasian | NHL | Related | Matched | 15 | 27 |
| Control 2 | 64 | Male | Caucasian | MDS | Unrelated | Matched | 27 | 49 |
| Case 3 | 54 | Female | Caucasian | MM | Unrelated | Matched | 21 | 30 |
| Control 3 | 57 | Female | Caucasian | NHL | Unrelated | Matched | 21 | |
| Case 4 | 53 | Female | Caucasian | MM | Related | Matched | 13 | |
| Control 4 | 57 | Female | Caucasian | MDS | Related | Matched | 20 | |
| Case 5 | 61 | Male | Caucasian | MM | Self | Matched | 16 | |
| Control 5 | 72 | Male | Caucasian | MM | Self | Matched | 16 | |
| Case 6 | 44 | Male | Caucasian | HD | Unrelated | Matched | 12 | 28 |
| Case 7 | 54 | Female | Caucasian | AML | Unrelated | Mismatchedc | 20 | 39 |
HLA, human leukocyte antigen; GVHD, graft-versus-host disease; AML, acute myelogenous leukemia; NHL, non-Hodgkin’s lymphoma; MDS, myelodysplastic syndrome; MM, multiple myeloma; HD, Hodgkin’s disease.
“Matched” indicates 10/10 allele or antigen matching.
Umbilical cord blood was used as the source of donor cells.
Number of days post-HCT.
Engraftment was defined as the first of three consecutive days of an absolute neutrophil count of >500 cells/µl.
GVHD was defined as previously described in reference 37.
mRNA isolation from PAXgene tubes.
We isolated sufficient mRNA to allow for high-throughput sequencing for five of seven cases and three of five controls (Table 3). There was substantial variability in complete blood counts for each subject at the time of PAXgene tube collection due to sample acquisition within the first few weeks after HCT (Table 3). The lowest yields of total RNA were observed for individuals with the lowest absolute white blood cell (WBC) counts. The two cases and two controls with insufficient RNA recovery had PAXgene tubes obtained the earliest after HCT (within 14 days) and prior to neutrophil engraftment.
TABLE 3.
Total WBC count (for whole-blood samples), mRNA isolation, and HHV-6B gene transcript counts in post-HCT whole-blood samples, DLBCL tissue samples, and SupT1 CD4+ T cells with lytic HHV-6B Z29 infectiona
| Sample type | Total WBCb (cells/mm3) | Total mRNA (ng) | Total HHV-6B VPMM |
|---|---|---|---|
| Whole blood | |||
| Case 1 | 7.0 | 5,075 | 0.02c |
| Case 2 | 5.5 | 2,523 | 14 |
| Case 3 | 2.1 | 6,687 | 0.03c |
| Case 4 | 3.7 | 9,529 | 0 |
| Case 5 | 0.06 | Insufficient | ND |
| Case 6 | 1.6 | Insufficient | ND |
| Case 7 | 5.2 | 3,973 | 19 |
| Control 1 | 3.0 | 9,222 | 0 |
| Control 2 | 1.3 | 6,467 | 0 |
| Control 3 | 0.3 | Insufficient | ND |
| Control 4 | 9.3 | 12,093 | 0 |
| Control 5 | 0.2 | Insufficient | ND |
| DLBCL tissue | |||
| Case 1 | NA | ND | 21 |
| Case 2 | NA | ND | 94 |
| SupT1 CD4+ T cells | NA | ND | 7,040 |
WBC, white blood cell; VPMM, viral reads per million unique mapped genomic reads; DLBCL, diffuse large B cell lymphoma; NA, not applicable; ND, not done.
WBC counts were drawn on the same day as the PAXgene tube.
U38 was the only transcript detected.
Comparison of the HHV-6B transcriptome in whole blood, DLBCL tumor tissue, and SupT1 CD4+ T cells with lytic HHV-6B Z29 infection.
We sought to characterize and compare the relative abundances of HHV-6B transcripts between whole blood from post-HCT patients with and without HHV-6B plasma viremia, tissue samples from patients with DLBCL, and a SupT1 CD4+ T cell culture with lytic HHV-6B Z29 infection. In post-HCT whole-blood samples from the five cases and three controls with sufficient RNA isolation, transcriptome analysis using RNA-seq identified HHV-6B gene expression in four of five cases and zero of three controls (Table 3). There was no evidence of correlation between plasma HHV-6B DNA viral loads detected by qPCR (Table 1) and the total HHV-6B gene transcript counts in whole-blood samples detected by RNA-seq (Table 3), although this comparison was limited by differential times between sample collection, differential total mRNA isolation given variability in sample cellularity, and the small sample size. Overall, whole-blood samples had low HHV-6B gene counts, with a range of 0.03 to 19 viral reads per million unique mapped genomic reads (VPMM) per sample (Table 3). In comparison, tumor tissue samples from two patients with DLBCL had 21 and 94 HHV-6B VPMM (Table 3). The in vitro SupT1 CD4+ T cell culture with lytic HHV-6B Z29 infection had the highest HHV-6B gene count, with 7,040 VPPM (Table 3).
In post-HCT whole blood, a broad array of HHV-6B genes was detected from cases 2 and 7, including immediate early, early, and late genes (Fig. 1). In cases 1 and 3, we only detected HHV-6B reads mapping to ORF U38. Tumor tissue samples from the two patients with DLBCL had an overall increased diversity of HHV-6B gene expression for most HHV-6B genes. The in vitro SupT1 CD4+ T cell culture with lytic HHV-6B infection demonstrated the greatest diversity of HHV-6 gene expression. Transcripts for U13 and the B genes (B1, B2, B3, B4, B5, B8, and B9) were detected only in the HHV-6B-infected SupT1 CD4+ T cell culture. The complete data sets for absolute, normalized, and relative HHV-6B gene transcript count detection are shown in Tables S1 to S3, respectively.
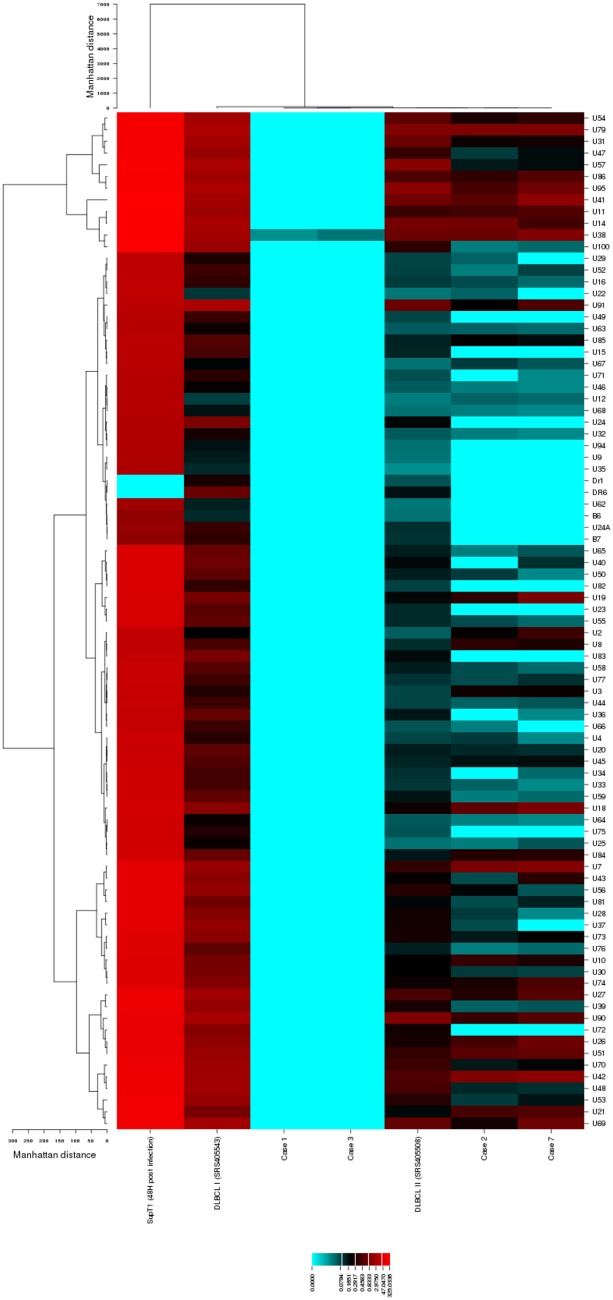
FIG 1.
Heat map of hierarchical clustering analysis of the HHV-6B transcriptome from whole blood, DLBCL tumor tissue, and SupT1 CD4+ T cells with lytic HHV-6B Z29 infection. We performed hierarchical clustering analysis to compare the normalized read numbers mapped to each HHV-6B gene in whole blood, DLBCL tumor tissue, and SupT1 CD4+ T cells with lytic HHV-6B Z29 infection (48 h postinfection). Hierarchically clustered viral genes (rows) from each sample type (columns) are shown in dendrograms and demonstrate differential clustering. Cases 1, 2, 3, and 7 are from whole-blood samples in subjects with HHV-6B DNA detection in plasma after allogeneic HCT. DLBCLs I and II are tumor tissue samples from two individuals with DLBCL. DLBCL I was coinfected with Epstein-Barr virus.
We performed hierarchical clustering analysis to compare the normalized read numbers mapped to each HHV-6B gene from whole-blood samples, tumor tissue samples, and the SupT1 CD4+ T cell culture with lytic HHV-6B infection at 48 h postinfection. The results are displayed in the form of a heat map in Fig. 1. In this analysis, whole-blood samples clustered together with one case of DLBCL. The other case of DLBCL clustered separately, indicating a distinct viral gene expression pattern. The SupT1 CD4+ T cell culture had exponentially higher gene expression across the HHV-6B genome and also clustered separately.
We compared the top ten HHV-6B transcripts with the highest normalized number of reads from each sample type (Table 4). This demonstrated that reads mapping to HHV-6B transcripts associated with expression of ORF U38, a viral polymerase gene, were the only transcripts detected in all the RNA-seq data sets. Overall, five of the top 10 HHV-6B genes (ORFs U95, U79, U41, U38, and U14) were shared between all three sample types, demonstrating high concordance between in vitro and in vivo HHV-6B gene transcripts in the setting of lytic infection. Reads mapping to late gene transcripts (ORFs U57, U54, U41, and U38) were more prevalent in the DLBCL tissue samples than whole-blood samples (ORF U38 only).
TABLE 4.
The 10 most highly expressed HHV-6B gene transcripts detected in post-HCT whole-blood samples, DLBCL tissue samples, and SupT1 CD4+ T cells with lytic HHV-6B Z29 infectiona
| Whole blood (n = 2)b |
DLBCL (n = 2) |
SupT1 CD4+ T cells (n = 1) |
|||
|---|---|---|---|---|---|
| Gene | Avg VPMMc | Gene | Avg VPMMc | Gene | VPMM |
| U42 (E) | 0.5 | U95 (IE) | 2.4 | U41 (E) | 325.0 |
| U7 (E) | 0.5 | U57 (L) | 2.3 | U11 (L) | 286.0 |
| U41 (E) | 0.5 | U79 (IE) | 2.1 | U14 (E) | 285.9 |
| U38 (L) | 0.4 | U90 (E) | 1.8 | U38 (L) | 269.4 |
| U79 (IE) | 0.4 | U14 (E) | 1.7 | U100 (L) | 264.2 |
| U18 (E) | 0.4 | U91 (E) | 1.7 | U57 (L) | 243.4 |
| U95 (IE) | 0.3 | U41 (E) | 1.5 | U86 (IE) | 222.3 |
| U14 (E) | 0.3 | U31 (L) | 1.4 | U95 (IE) | 222.1 |
| U51 (E) | 0.3 | U38 (L) | 1.4 | U54 (L) | 211.5 |
| U19 (E) | 0.3 | U54 (L) | 1.3 | U79 (IE) | 207.7 |
Genes in bold are those that were identified in the10 most highly expressed transcripts for all sample types. We used only one SupT1 CD4+ T cell culture. The temporal pattern of expression is defined as previously described (38). E, early; IE, immediate early; L, late.
The data sets obtained from cases 1 and 3 were omitted due to a low number of reads to only one transcript (U38).
Normalized read counts expressed in VPMM were averaged between samples.
HHV-6B transcriptome in B-LCLs with latent HHV-6B infection and iciHHV-6B.
We did not detect reads aligning to any specific HHV-6B transcripts in data sets from a beta lymphoblastoid cell line (B-LCL) sample with latent HHV-6B infection from postnatal infection or a B-LCL sample with iciHHV-6B.
Development and validation of an RT-qPCR assay for ORF U38.
We selected ORF U38 as the best candidate for a diagnostic target to identify lytic HHV-6B replication in biologic samples based on our findings that ORF U38 was expressed in all sample types and the only transcript detected in all samples with HHV-6B gene expression. To confirm the detection of reads mapping to ORF U38, we developed a specific RT-qPCR assay to detect this mRNA transcript. Using this newly developed assay, we tested mRNA extracted from whole blood in four of the post-HCT cases with HHV-6B plasma detection who had a second available PAXgene tube. We detected the ORF U38 transcript in all four cases (Table 5). Additionally, the high sensitivity of this assay was demonstrated by the detection of U38 mRNA from one subject in whom RNA-seq did not identify any HHV-6B gene transcripts (case 4), as well as another case in which there was an insufficient amount of isolated RNA to perform RNA-seq (case 5). The number of ORF U38 copies detected by RT-qPCR correlated with the number of reads mapping to ORF U38 by RNA-seq (r2 = 1.0; P = 0.08), but this did not reach statistical significance, likely due to the small sample size.
TABLE 5.
HHV-6B ORF U38 detection by RNA-seq and RT-qPCR in post-HCT whole-blood samples
| Subject | Sample description | U38 reads, RNA-seqa | U38 RNA copies/ml, RT-qPCR | U38 DNA copies/ml, qPCRb |
|---|---|---|---|---|
| Case 3 | Whole blood | 8 | 19c | 0 |
| Case 4 | Whole blood | 0 | 754 | 0 |
| Case 5 | Whole blood | NDd | 78 | 0 |
| Case 7 | Whole blood | 102 | 106,188 | 213 |
| Control 1 | Whole blood | 0 | 0 | 0 |
| Control 2 | Whole blood | 0 | 0 | 0 |
| Control 3 | Whole blood | 0 | 0 | 0 |
| Control 4 | Whole blood | 0 | 0 | 0 |
| Control 5 | Whole blood | 0 | 0 | 0 |
| B-LCL 1 | Postnatal HHV-6B infection | 0 | 0 | 0 |
| B-LCL 2 | iciHHV-6B | 0 | 30.4e | 30.1e |
Absolute read count (from Table S1).
DNA contamination was assessed by testing the RNA samples without performing a reverse transcription step to identify background cDNA.
This value corresponds to a detection level of approximately 6 copies per reaction, which is within the detection limit of the assay.
There was insufficient RNA isolated to allow for RNA-seq.
This result is expressed as the cycle threshold, not copies per milliliter. The cycle threshold values of the assays with and without reverse transcriptase (to demonstrate U38 RNA and DNA, respectively) were identical, which indicated that the U38 signal was from residual contaminating DNA.
To determine the specificity of the RT-qPCR assay for U38, we tested mRNA extracted from whole blood from all 5 post-HCT controls without HHV-6B DNA plasma detection, as well as the 2 B-LCLs with HHV-6 DNA detection but no HHV-6B gene transcript reads identified by RNA-seq. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was identified by RT-qPCR in all samples, demonstrating sufficient isolation of RNA. However, U38 mRNA was not detected in any of these samples from subjects without evidence of HHV-6B replication, supporting the specificity the RT-qPCR assay for U38 (Table 5).
Because the HHV-6B ORF U38 is not a spliced gene product, there is a possibility of amplification of contaminating HHV-6B genomic DNA. Although we performed a DNase treatment to mitigate contaminating DNA, we also performed an RT-qPCR excluding reverse transcriptase to assess for genomic HHV-6B DNA contamination. There was evidence of minimal DNA contamination in only one sample (case 7 [Table 5]).
Host cellular gene transcriptome and cytokines in whole-blood samples from post-HCT cases and controls.
We performed a differential gene expression analysis of host cellular gene expression profiles in whole blood comparing normalized read counts from RNA-seq data sets between post-HCT cases and controls to determine whether we could distinguish between individuals with and without HHV-6B reactivation. We did not observe statistically significant differences in cellular gene expression in any of the paired samples (data not shown). Using a commercially available Luminex-based assay, we also tested plasma samples obtained pre- and post-PAXgene tube whole-blood collection for 14 immunoinflammatory cytokines from cases and controls. All the cytokines tested in plasma were at or below the limit of detection and precluded analysis of differential cytokine expression between individuals with and without HHV-6B reactivation (data not shown).
Structure of selected HHV-6B transcripts from whole-blood and DLBCL tumor tissue samples during HHV-6B replication.
We recently reported differential splicing patterns of several HHV-6B transcripts in cell lines following in vitro infection, sometimes involving unpredicted splicing sites compared to the reference genome annotation (21). These findings could affect the performance of diagnostic RT-qPCR assays targeting these transcripts. In the current study, we characterized selected HHV-6B spliceoforms that were detected at relatively high levels (ORFs U79, U95, and U100) in post-HCT whole-blood and DLBCL tumor tissue samples. Our analysis of the mapped reads revealed that all detected splice sites contained the canonical dinucleotides GT and AG for donor and acceptor sites, respectively (Table 6).
TABLE 6.
Donor and acceptor sites for spliced HHV-6B gene transcriptsa
| Gene | Donor sequence | Acceptor sequence |
|---|---|---|
| U79 | ||
| Exon I | AATTTGATGGTAAA | ATTTCAGAACCCCC |
| Exon II (short) | CGGTTGAGGGTGCG | CTTTTAGGGGACG |
| Exon II (long) | TATGCGCAGGTGGG | CTTTTAGGGGACG |
| U95 | CGGTACAGGGTGAG | CACCTCTAGATGTCT |
| U100 or Q1/Q2 | ||
| Exon | GGCCACGGTAGG | TTTTCGCAAGACTAAG |
| Exon I (ATG) | NA | NA |
| Exon II | GGCTCGATGGTGAG | AAAATCCAGCGAACT |
| Exon III | CTACGTCGTGTG | TTTCGTCAGATTTCA |
| Exon IV | TTCTGTACTGTAAG | TTCTCACAGAGCGTT |
| Exon V | ATCTATACGGTGAG | TATGCACAGCAACAT |
| Exon VI | CGCATTATGGTGAG | AATCCGCAGCACCAT |
| Exon VII | NA | NA |
| Exon VIII (short) | CGGTGAAATGTAAG | TTTGTTCAGTCTGTT |
| Exon VIII (long) | CGGTGAAATGTAAG | AAATGCAGGCCAGA |
| Exon IX | NA | NA |
| Exon X | AGTCATTAGGTATT | TTTTTTAAGTGACTA |
Donor and acceptor splice site consensus sequences are in bold. “NA” indicates that there were no reads mapping to that region.
For the HHV-6B U79 transcript, four different mRNA spliceoforms have been previously described and sequenced (22). An additional U79 mRNA isoform was recently identified in T cell lines following experimental infection with HHV-6B, and its translation into protein was confirmed using mass spectrometry (21). In our current study, RNA-seq data from whole-blood samples demonstrated an additional novel U79 splice variant corresponding to an open reading frame including exon I, exon II, intron II, and a partial sequence from exon III (Fig. 2). Using proteomic data from a previously published study (21), we discovered an HHV-6B peptide sequence corresponding to the sequence encoded by intron II of the U79 mRNA (peptide sequence VYAQVGGVLGSPKR). This U79 mRNA isoform was also detected in one of the 3 U79 mRNA isoforms from the DLBCL tumor tissue.
FIG 2.
Novel splice variant of the HHV-6B U79 transcript. The RNA-seq data from whole-blood samples suggested a novel splice variant of the HHV-6B U79 transcript compatible with a mRNA sequence corresponding to an open reading frame including exon I, exon II, intron II, and a partial sequence from exon III. This U79 mRNA isoform was also detected in one of the three U79 mRNA isoforms identified in the RNA-seq data sets from DLBCL tumor samples.
The structure of the HHV-6B U95 transcript has been previously reported (22). Our RNA-seq data from whole-blood samples confirmed that the transcription start site is located upstream of the initiating ATG sequence and that the transcript consists of two exons on both sides of ORF U94.
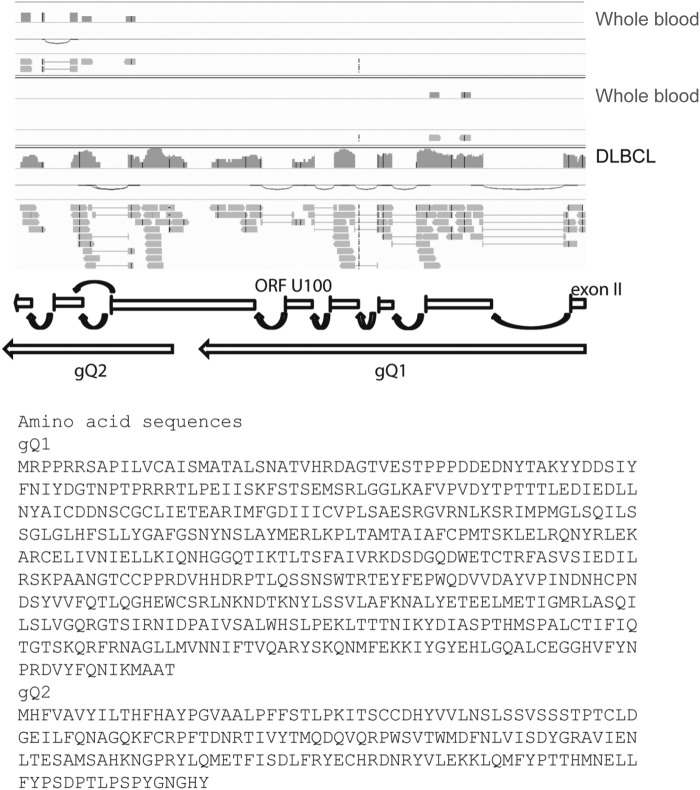
For the HHV-6B U100 transcript, a previous study showed the viral transcripts encode two glycoproteins designated gQ1 and gQ2, both of which are expressed on the viral envelope in the form of a tetrameric complex with glycoproteins gH and gL (22). Our RNA-seq data from whole-blood samples revealed reads mapping to either gQ1 or gQ2. In contrast, data from the DLBCL tumor tissue reads mapped to both gQ1 and gQ2 simultaneously. These observations suggest that expression of each glycoprotein could be independently regulated, which is further supported by the existence of a poly(A) sequence at the end of the gQ1 open reading frame. Additionally, our analysis of reads mapping to the gQ2 transcript showed two possible splice sites, and the mature transcript corresponding to the alternative gQ2 splice is predicted to code for a shorter protein (68 amino acids versus 182 amino acids [Fig. 3]). Further experiments are required to establish the biological relevance of this gQ2 protein isoform.
FIG 3.
Two possible splice sites for the HHV-6B U100 gQ2 transcript. RNA-seq HHV-6B gene transcript reads from whole blood and DLBCL tumor samples mapping to the U100 gQ2 transcript showed two possible splice sites. Both predicted intron sequences have typical splicing donor and acceptor sites (Table 6). The mature transcript corresponding to the alternative gQ2 splice is predicted to code for a shorter protein (68 amino acids versus 182 amino acids).
DISCUSSION
We used RNA-seq to characterize the in vivo HHV-6B transcriptome in whole-blood samples collected from allogeneic HCT recipients and compared this to the HHV-6B transcriptome in DLBCL tumor tissue and to HHV-6B-infected SupT1 CD4+ cells in culture. We observed concordance between the HHV-6B transcriptome observed in the in vivo and in vitro samples, although there was substantial variability in the breadth and quantity of gene expression across sample types. The HHV-6B viral polymerase gene U38 was the only transcript detected in all RNA-seq data sets and was one of the top 10 most highly transcribed genes. Given the relatively low abundance of reads mapping to U38 by RNA-seq, we developed a novel RT-qPCR assay for U38 to employ an orthogonal method to obtain an accurate absolute measurement for the presence of the U38 transcript. Using this assay, we detected the HHV-6B U38 gene in all tested whole-blood samples from patients with concurrent HHV-6B viremia, suggesting that this is a sensitive assay for HHV-6B replication. We also demonstrated discrepancies with the predicted splice sites for the HHV-6B U79 and U100 genes in whole-blood samples compared to the HHV-6B reference genome, so caution is required if designing PCR assays for these targets.
Quantitative PCR to detect HHV-6B genomic DNA is the standard clinical approach for diagnostic testing. However, this has important limitations in specificity given that most humans are infected with HHV-6B early in life, HHV-6B remains latent in a diverse array of cell types, and about 1% of the population has iciHHV-6 (species A or B) with a high burden of cell-associated HHV-6 DNA (10). Thus, it may be difficult to distinguish between latent and actively replicating virus when HHV-6B DNA is detected by qPCR in cellular samples. For a related herpesvirus, cytomegalovirus (CMV), the use of RT-qPCR to identify CMV transcripts in bronchoalveolar lavage fluid of immunocompromised patients demonstrated significantly greater specificity for CMV pneumonia than standard qPCR, which has poor specificity due to the detection of latent virus and viral shedding (23). Development of RT-qPCR assays to detect HHV-6B mRNA may provide a more accurate method for identifying active infection, and studies using RT-qPCR to detect spliced transcripts have demonstrated relatively high sensitivity and specificity for viral replication in vivo (11, 16, 17). However, HHV-6B transcript targets that have been studied to date were selected based on their temporal expression pattern or because they were a spliced gene product.
This is the first study to our knowledge using RNA-seq to identify the HHV6-B gene expression profile in whole blood collected during periods of HHV-6B reactivation. Previous reports have characterized the expression pattern of several HHV-6B transcripts following experimental in vitro infection of different T cell lines with HHV-6B (21), and one study described the HHV-6B transcriptome in tumor tissue samples from two patients with DLBCL (19), although the pathogenic role of HHV-6B in this context is unclear. RNA-seq for transcriptome analysis provides an unbiased approach to identify consistently and highly expressed genes that may increase the sensitivity of targeted RT-qPCR assays for HHV-6B replication in biologic samples. Given that total mRNA isolation from peripheral blood mononuclear cells (PBMCs) in blood samples may be limited by low yields, degradation, coagulation, and altered gene transcription resulting from cell purification procedures (24), we chose to perform direct mRNA isolation from whole-blood samples using PAXgene tubes to allow for immediate and long-term stabilization of intracellular RNA (24).
We used RNA-seq to demonstrate a broad range of viral gene expression across the HHV-6B genome in whole blood from subjects with concurrent plasma viremia, indicative of lytic replication. We did not detect any HHV-6B transcripts in subjects without concurrent plasma viremia. We also did not detect HHV-6B transcripts in B-LCL samples with latent HHV-6B, although we identified reads containing the TAACCC nucleotide sequence. We could not determine the origin of this transcript given that this telomeric repeat sequence is found in both HHV-6B and human genomes (25). Interestingly, reads containing the TAACCC sequence were not systematically detected in all the samples analyzed in our study. This observation suggests that some of these reads could originate from transcription of the telomeric repeat region of the viral genome.
The previously reported study of two DLBCL tumor samples with HHV-6B infection demonstrated higher RNA-seq read counts mapping to the HHV-6B genome with broader gene expression profiles, including almost all known HHV-6B genes (19). Hierarchical cluster analysis of the HHV-6B transcriptome from whole-blood versus DLBCL tumor samples with HHV-6B infection showed similarities but also differences in gene expression profiles that may be explained in part by differences in infected cell types. An additional source of differential gene expression across samples and sample types may be the timing of sampling relative to viral reactivation. Studies have demonstrated differences in the abundances of HHV-6B genes, including U38, at different time points during the lytic cycle after initial viral infection (26, 27). In this study, we were unable to formally assess temporal HHV-6B gene expression patterns given our lack of longitudinally obtained samples. Nonetheless, we identified five gene transcripts that were represented among the top 10 most highly enumerated HHV-6B transcripts in in vivo and in vitro samples (Table 4).
HHV-6B U38 was the only gene transcript detected in all the RNA-seq data sets and was the only viral transcript detectable in two of the whole-blood samples. Interestingly, HHV-6B U38 appears to be an immunodominant epitope recognized by human CD4+ T cells (28). We developed a targeted RT-qPCR assay for U38. This assay detected HHV-6B U38 in all tested whole-blood samples from subjects with HHV-6 viremia and in no whole-blood samples from subjects without HHV-6 viremia, demonstrating the potential utility of this assay for identifying HHV-6B replication in clinical samples. Although U38 is not a spliced gene product leading to the possibility of contamination with genomic DNA, we demonstrated minimal contamination after treatment with DNase. A recent study reported detection of a small noncoding RNA for U14 in multiple sample types from a patient with HHV-6A reactivation (27). U14 was also one of the most frequently detected transcripts in our cohort, supporting the potential of U14 as an additional diagnostic target. Whereas prior studies have tested the sensitivity and specificity of assays targeting HHV-6B transcripts such as U12, U16/17, U60/66, U79/U80, U89/90, and U100 (11–17), U79 was the only one of these transcripts shared across sample types at a relatively high abundance. Our data also demonstrated potential unique spliceoforms for U79 and U100, which may further compromise the sensitivity of PCR assays for these targets.
We also used the RNA-seq data from whole blood to study whether unique host gene expression or cytokine profiles could be demonstrated among patients with and without HHV-6 reactivation post-HCT. Provocative data using microarrays demonstrated a unique host gene expression profile in children with primary HHV-6B infection (26), and other studies have shown associations between specific cytokine levels and HHV-6 infection after HCT (29). However, we did not find any significant differences in whole-blood host gene expression and cytokine profiles between subjects with and without HHV-6 reactivation in this study. However, our sample size was small, no subjects in our cohort had severe HHV-6 infection with end-organ disease, and biologic changes early after HCT (e.g., donor cell engraftment and graft-versus-host disease) likely had larger effects on host gene expression.
Strengths of this study included the use of RNA-seq analyses of unique biologic samples to characterize in vivo HHV-6B gene expression and comparison of these findings with in vitro HHV-6B gene expression. Furthermore, we used these data to generate an evidence-based RT-qPCR assay to identify HHV-6B replication in biologic samples. The low overall number of HHV-6B gene transcript reads prevented us from accurately determining the relative abundance of each transcript. Nonetheless, our characterization of the HHV-6B transcriptome in HCT patients with HHV-6B reactivation provides a comprehensive list of viral proteins of biological interest. Future studies using sequentially collected samples from subjects before and after HHV-6B reactivation, as well as from subjects with iciHHV-6, will be important to refine these findings.
In conclusion, we demonstrated that RNA-seq can be used to determine HHV-6B gene expression after allogeneic HCT, and these analyses identified the HHV-6B U38 mRNA transcript as a potential target for focused clinical diagnostic assays such as RT-qPCR. We developed a novel RT-qPCR assay targeting HHV-6B U38 and detected U38 mRNA in all tested samples from subjects with HHV-6B viremia, underscoring the potential for this biomarker to identify active viral replication in clinical samples. This assay may be particularly useful to identify lytic HHV-6B infection in nonplasma samples and samples from individuals with iciHHV-6B. Larger studies that correlate HHV-6B mRNA transcript detection with HHV-6B genomic DNA detection and HHV-6B disease will be critical to establish actionable DNA and mRNA transcript thresholds for treatment.
MATERIALS AND METHODS
Patients.
This study was approved by the Fred Hutchinson Cancer Research Center Institutional Review Board. We identified a cohort of patients who had whole blood collected in PAXgene blood RNA tubes (PAXgene tubes; Qiagen, Germantown, MD) within the first 6 weeks of autologous or allogeneic HCT as part of an unrelated study. We then identified plasma samples from these subjects that were collected on the day of PAXgene tube collection and temporally proximal, both before and after. We tested these samples for HHV-6B DNA using qPCR. Patients who had plasma HHV-6B DNA detected within 7 days of PAXgene tube collection were selected for this study and were categorized as cases; one patient with 11 days between sample collection and HHV-6B detection was included (Table 1). Patients without HHV-6B detection in plasma samples within 7 days of PAXgene tube acquisition were used as controls and matched in a 1-to-1 ratio to cases based on the day post-HCT of PAXgene tube collection, age, race, sex, type of HCT (autologous versus allogeneic), donor relation, human leukocyte antigen (HLA) matching, neutrophil engraftment status, and acute graft-versus-host disease (aGVHD) status (Table 1). We used plasma samples to identify patients with lytic versus latent HHV-6B infections as a model system given the high sensitivity and specificity of plasma HHV-6B DNA detection for lytic HHV-6B infection (30).
We also used data from two patients with DLBCL from a published study in which RNA-seq on tumor tissue samples demonstrated a broad profile of HHV-6B transcripts consistent with lytic transcription (19).
Samples.
PAXgene tubes were filled with 2.5 ml of whole blood obtained directly from HCT recipients, and samples were stored according to the manufacturer’s instructions. Plasma samples were leftover clinical specimens used for cytomegalovirus testing. For in vitro comparisons, we obtained a cell culture of HHV-6B Z29-infected SupT1 CD4+ T cells (provided by the HHV-6 Foundation). Cells were harvested 48 h after infection.
We also obtained two beta lymphoblastoid cell lines (B-LCLs) with latent HHV-6B infection derived from pre-HCT patient PBMCs. One sample had low-level HHV-6B DNA detection likely due to postnatal HHV-6B infection with latency in lymphocytes. The other sample had high-level HHV-6B DNA detection (1 copy per cell) consistent with iciHHV-6B. These samples were identified in a prior study of iciHHV-6B, and the presence or absence of iciHHV-6B was tested by droplet digital PCR as previously described (9, 31).
HHV-6B DNA testing.
We tested plasma samples for HHV-6B DNA using high-throughput real-time qPCR that detects 1 copy of HHV-6 DNA per reaction (25 copies/ml) and distinguishes between species A and B as previously described (32).
RNA extraction for RNA sequencing.
We isolated and extracted total RNA from PAXgene tubes according to the manufacturer’s instructions using the MagMAX blood RNA kit (Ambion, Foster City, CA). RNA quality was determined by electropherogram (showing 28S, 18S, and 5S bands) and RNA integrity number using an Agilent 2200 bioanalyzer (Agilent Technologies, Santa Clara, CA). Total RNA concentration was obtained from an absorbance ratio at 260 and 280 nm using a NanoDrop ND-2000 spectrometry instrument (NanoDrop Technologies, Wilmington, DE). Since whole-blood RNA consists mostly of globin mRNA, we performed globin depletion using a GLOBINclear kit (Ambion) to maximize detection of less abundant mRNA and further purified and concentrated RNA on appropriate columns before RNA library preparation.
RNA sequencing.
We performed poly(A) RNA (coding mRNA) selection to minimize DNA contamination. RNA-seq libraries were prepared using the Ovation single-cell RNA-seq system (NuGen Technologies, Inc., San Carlos, CA). Library size distributions were validated using an Agilent 2200 TapeStation (Agilent Technologies, Santa Clara, CA). Additional library quality control, blending of pooled indexed libraries, and cluster optimization were performed using Life Technologies’ Invitrogen Qubit 2.0 fluorometer (Life Technologies-Invitrogen, Carlsbad, CA). Barcoded RNA-seq libraries were run individually on a flow cell lane using an Illumina cBot for whole blood from samples HCT recipients with HHV-6B viremia (cases) and the SupT1 CD4+ cell culture; whole blood from HCT recipients without HHV-6B viremia (controls) and B-LCLs were duplexed or triplexed. Sequencing was performed using an Illumina HiSeq 2500 in rapid mode employing a paired-end, 50-base read length sequencing strategy. Image analysis and base calling were performed using Illumina’s Real Time Analysis v1.18 software, followed by “demultiplexing” of indexed reads and generation of FASTQ files using Illumina’s bcl2fastq conversion software (v1.8.4). Reads that passed the quality check were aligned to a HHV-6B reference genome (GenBank accession number NC_000898) using TopHat (v2.09) along with Bowtie 2 (v2.2.3) (33, 34). Aligned reads obtained from TopHat were visualized using Geneious software (Biomatters Limited) to assess the extent and depth of coverage, sequence alignments, and identified splice junctions. Read counts per HHV-6B gene enumeration were generated using HTseq-count (v0.6.1), employing the “intersection-strict” overlap mode (35).
ORF U38 RT-qPCR assay development and testing.
We used primers (forward, TGCCCGATTYTGAAAAAGCT, and reverse, CCTGTGGGTATTCATAAAATTTTG) and probe (6-carboxyfluorescein [FAM]-CTCCCGCGCTTTGCACAGACG-black hole quencher [BHQ]) specific to a conserved region of ORF U38 using Primer Express 3.0.1 (Applied Biosystems, Foster City, CA). The assay was verified using standards created from primer-specific ORF U38 transcripts isolated from an HHV-6B-infected cell line. The lowest point on the standard curve for quantitation was 10 copies per reaction (approximately 30 copies per ml). For whole-blood samples, we extracted RNA from 2.5 ml of whole blood collected in PAXgene tubes using the PAXgene blood RNA kit (Qiagen, Germantown, MD) and eluted into 80 µl of BR5 buffer. For B-LCLs, we washed samples with 1× phosphate-buffered saline and extracted RNA using an RNeasy Plus kit (Qiagen). To prevent amplification of HHV-6B genomic DNA, all samples were treated with DNase as part of the extraction protocol. To determine the extent of RNA recovery, we tested samples for GAPDH RNA (Thermo Fisher Scientific, Waltham, MA). One step RT-PCR was performed using a QuantiTect probe RT-PCR kit (Qiagen) and the 7500 fast real-time PCR system. Each 25-µl RT-qPCR mixture contained 0.25 µl of QuantiTect virus RT mix (100×), 5 µl of QuantiTect virus master mix (5×), 0.5 µl of ROX reference dye (Thermo Fisher Scientific, Waltham, MA), 0.4 µM primers, 0.2 µM probe, 10 µl of RNA, and water to volume. The thermocycling conditions were as follows: 20 min at 50°C for reverse transcription, 5 min at 95°C for PCR activation, and 45 cycles at 95°C for 15 s followed by 45 s at 60°C. An RT-qPCR excluding reverse transcriptase was performed on all standards and samples to assess for DNA contamination.
Cytokine testing.
We selected plasma samples from three time points prior to, closest to, and after the day of PAXgene tube collection for cytokine immunoassay testing. We used a Luminex 200 system for multiplex microbead cytokine analyses, run in duplicate, to measure a panel of 14 immunoinflammatory cytokines, including interleukin 7 (IL-7), IL-9, IL-10, IL-15, IL-17, IL-21, IL-22, IL-23, IL-27, IL-31, IL-1α, IL-1RA, tumor necrosis factor beta (TNF-β), and alpha interferon (IFN-α).
Statistics.
To assess global differences in HHV-6B gene expression profiles from different sample types, we performed hierarchical clustering analysis using Clustered Image Maps software (CIMminer). The Manhattan distance matrix was computed based on the respective abundance of each viral gene detected in each patient. To allow comparisons between data sets obtained from different sample types, read counts per HHV-6B gene were normalized by calculating viral reads per million unique mapped genomic reads (VPMM). These values were used as input for hierarchical clustering using the complete linkage-clustering algorithm. Data sets representing viral gene expression profiles were visualized by heat maps.
We performed a differential gene expression analysis of host cellular gene expression profiles comparing normalized read counts from RNA-seq data sets between cases and controls. Paired analysis was performed using the Bioconductor package DESeq22 software (36).
Accession number(s).
RNA-seq generated from our samples are available at the Gene Expression Omnibus (GEO) website (accession number GSE115533). RNA-seq data sets for two patients with DLBCL (SRS405408 and SRS405443) were downloaded from the NIH database of genotypes and phenotypes using accession code phs000235.v2.p1. These RNA-seq data sets are freely available through controlled access (https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000235.v2.p1).
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge the Fred Hutchinson Cancer Research Center Research Cell Bank for providing B-LCL samples and the HHV-6 Foundation for providing HHV-6B-infected SupT1 CD4+ T cells. We thank the Cancer Genome Characterization Initiative for acquiring and sequencing the DLBCL samples analyzed in this study. We also thank Terry Stevens-Ayers, Elsa Garnace, and Zach Stednick for help obtaining samples and data.
This work was supported by the National Institutes of Health (5K23AI119133-03 to J.A.H. and K24HL093294 to M.B.). Additional resources were provided by the National Institutes of Health (HL088021, CA78902, CA18029, CA15704, HL122173, and P50 HL110787) and the Fred Hutch Vaccine and Infectious Disease Division (support of the sample biorepository).
All authors declare no conflicts of interests related to the manuscript.
J.A.H., M.B., and S.B. were responsible for the design of the study. J.A.H., M.I., R.S.B., V.P., M.-L.H., R.H.S., K.R.J., and S.B. analyzed the data. Samples and data were collected by J.A.H. and S.B. J.A.H., D.M.Z., R.S.B., R.H.S., K.R.J., M.B., and S.B. interpreted the data. J.A.H. and S.B. wrote the first draft. All authors contributed to the writing and revision of the manuscript.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JVI.01419-18.
REFERENCES
- 1.Hill JA, Zerr DM. 2014. Roseoloviruses in transplant recipients: clinical consequences and prospects for treatment and prevention trials. Curr Opin Virol 9:53–60. doi: 10.1016/j.coviro.2014.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Scheurer ME, Pritchett JC, Amirian ES, Zemke NR, Lusso P, Ljungman P. 2013. HHV-6 encephalitis in umbilical cord blood transplantation: a systematic review and meta-analysis. Bone Marrow Transplant 48:574–580. doi: 10.1038/bmt.2012.180. [DOI] [PubMed] [Google Scholar]
- 3.Achour A, Boutolleau D, Slim A, Agut H, Gautheret-Dejean A. 2007. Human herpesvirus-6 (HHV-6) DNA in plasma reflects the presence of infected blood cells rather than circulating viral particles. J Clin Virol 38:280–285. doi: 10.1016/j.jcv.2006.12.019. [DOI] [PubMed] [Google Scholar]
- 4.Hill JA, Boeckh MJ, Sedlak RH, Jerome KR, Zerr DM. 2014. Human herpesvirus 6 can be detected in cerebrospinal fluid without associated symptoms after allogeneic hematopoietic cell transplantation. J Clin Virol 61:289–292. doi: 10.1016/j.jcv.2014.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hill JA, Koo S, Guzman Suarez BB, Ho VT, Cutler C, Koreth J, Armand P, Alyea EP, Baden LR, Antin JH, Soiffer RJ, Marty FM. 2012. Cord-blood hematopoietic stem cell transplant confers an increased risk for human herpesvirus-6-associated acute limbic encephalitis: a cohort analysis. Biol Blood Marrow Transplant 18:1638–1648. doi: 10.1016/j.bbmt.2012.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fotheringham J, Akhyani N, Vortmeyer A, Donati D, Williams E, Oh U, Bishop M, Barrett J, Gea-Banacloche J, Jacobson S. 2007. Detection of active human herpesvirus-6 infection in the brain: correlation with polymerase chain reaction detection in cerebrospinal fluid. J Infect Dis 195:450–454. doi: 10.1086/510757. [DOI] [PubMed] [Google Scholar]
- 7.Nagate A, Ohyashiki JH, Kasuga I, Minemura K, Abe K, Yamamoto K, Ohyashiki K. 2001. Detection and quantification of human herpesvirus 6 genomes using bronchoalveolar lavage fluid in immunocompromised patients with interstitial pneumonia. Int J Mol Med 8:379. [DOI] [PubMed] [Google Scholar]
- 8.Kaufer BB, Flamand L. 2014. Chromosomally integrated HHV-6: impact on virus, cell and organismal biology. Curr Opin Virol 9:111–118. doi: 10.1016/j.coviro.2014.09.010. [DOI] [PubMed] [Google Scholar]
- 9.Hall Sedlak R, Hill JA, Nguyen T, Cho M, Levin G, Cook L, Huang M-L, Flamand L, Zerr DM, Boeckh M, Jerome KR. 2016. Detection of HHV-6B reactivation in hematopoietic cell transplant recipients with inherited chromosomally integrated HHV-6A by droplet digital PCR. J Clin Microbiol 54:1223–1227. doi: 10.1128/JCM.03275-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hill JA, Hall Sedlak R, Jerome KR. 2014. Past, present, and future perspectives on the diagnosis of roseolovirus infections. Curr Opin Virol 9:84–90. doi: 10.1016/j.coviro.2014.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Norton RA, Caserta MT, Hall CB, Schnabel K, Hocknell P, Dewhurst S. 1999. Detection of human herpesvirus 6 by reverse transcription-PCR. J Clin Microbiol 37:3672–3675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Van den Bosch G, Locatelli G, Geerts L, Fagà G, Ieven M, Goossens H, Bottiger D, Oberg B, Lusso P, Berneman ZN. 2001. Development of reverse transcriptase PCR assays for detection of active human herpesvirus 6 infection. J Clin Microbiol 39:2308–2310. doi: 10.1128/JCM.39.6.2308-2310.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kondo K, Kondo T, Shimada K, Amo K, Miyagawa H, Yamanishi K. 2002. Strong interaction between human herpesvirus 6 and peripheral blood monocytes/macrophages during acute infection. J Med Virol 67:364–369. doi: 10.1002/jmv.10082. [DOI] [PubMed] [Google Scholar]
- 14.Pradeau K, Bordessoule D, Szelag J-C, Rolle F, Ferrat P, Le Meur Y, Turlure P, Denis F, Ranger-Rogez S. 2006. A reverse transcription-nested PCR assay for HHV-6 mRNA early transcript detection after transplantation. J Virol Methods 134:41–47. doi: 10.1016/j.jviromet.2005.11.015. [DOI] [PubMed] [Google Scholar]
- 15.Strenger V, Caselli E, Lautenschlager I, Schwinger W, Aberle SW, Loginov R, Gentili V, Nacheva E, DiLuca D, Urban C. 2014. Detection of HHV-6-specific mRNA and antigens in PBMCs of individuals with chromosomally integrated HHV-6 (ciHHV-6). Clin Microbiol Infect 20:1027–1032. doi: 10.1111/1469-0691.12639. [DOI] [PubMed] [Google Scholar]
- 16.Ihira M, Enomoto Y, Kawamura Y, Nakai H, Sugata K, Asano Y, Tsuzuki M, Emi N, Goto T, Miyamura K, Matsumoto K, Kato K, Takahashi Y, Kojima S, Yoshikawa T. 2012. Development of quantitative RT-PCR assays for detection of three classes of HHV-6B gene transcripts. J Med Virol 84:1388–1395. doi: 10.1002/jmv.23350. [DOI] [PubMed] [Google Scholar]
- 17.Bressollette-Bodin C, Nguyen TVH, Illiaquer M, Besse B, Peltier C, Chevallier P, Imbert-Marcille B-M. 2014. Quantification of two viral transcripts by real time PCR to investigate human herpesvirus type 6 active infection. J Clin Virol 59:94–99. doi: 10.1016/j.jcv.2013.11.014. [DOI] [PubMed] [Google Scholar]
- 18.Tsao EH, Kellam P, Sin CSY, Rasaiyaah J, Griffiths PD, Clark DA. 2009. Microarray-based determination of the lytic cascade of human herpesvirus 6B. J Gen Virol 90:2581–2591. doi: 10.1099/vir.0.012815-0. [DOI] [PubMed] [Google Scholar]
- 19.Strong MJ, O'Grady T, Lin Z, Xu G, Baddoo M, Parsons C, Zhang K, Taylor CM, Flemington EK. 2013. Epstein-Barr virus and human herpesvirus 6 detection in a non-Hodgkin’s diffuse large B-cell lymphoma cohort by using RNA sequencing. J Virol 87:13059–13062. doi: 10.1128/JVI.02380-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hill JA, Ikoma M, Zerr DM, Basom RS, Peddu V, Huang M-L, Hall Sedlak R, Jerome KR, Boeckh M, Barcy S. 2018. RNA sequencing of the in vivo human herpesvirus 6B transcriptome to identify targets for clinical assays distinguishing between latent and active infections. bioRxiv 10.1101/397679. [DOI] [PMC free article] [PubMed]
- 21.Greninger AL, Knudsen GM, Roychoudhury P, Hanson DJ, Sedlak RH, Xie H, Guan J, Nguyen T, Peddu V, Boeckh M, Huang M-L, Cook L, Depledge DP, Zerr DM, Koelle DM, Gantt S, Yoshikawa T, Caserta M, Hill JA, Jerome KR. 2018. Comparative genomic, transcriptomic, and proteomic reannotation of human herpesvirus 6. BMC Genomics 19:204. doi: 10.1186/s12864-018-4604-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Taniguchi T, Shimamoto T, Isegawa Y, Kondo K, Yamanishi K. 2000. Structure of transcripts and proteins encoded by U79-80 of human herpesvirus 6 and its subcellular localization in infected cells. Virology 271:307–320. doi: 10.1006/viro.2000.0326. [DOI] [PubMed] [Google Scholar]
- 23.Boivin G, Olson CA, Quirk MR, Kringstad B, Hertz MI, Jordan MC. 1996. Quantitation of cytomegalovirus DNA and characterization of viral gene expression in bronchoalveolar cells of infected patients with and without pneumonitis. J Infect Dis 173:1304–1312. doi: 10.1093/infdis/173.6.1304. [DOI] [PubMed] [Google Scholar]
- 24.Rainen L, Oelmueller U, Jurgensen S, Wyrich R, Ballas C, Schram J, Herdman C, Bankaitis-Davis D, Nicholls N, Trollinger D, Tryon V. 2002. Stabilization of mRNA expression in whole blood samples. Clin Chem 48:1883–1890. [PubMed] [Google Scholar]
- 25.Strong MJ, Blanchard E, Lin Z, Morris CA, Baddoo M, Taylor CM, Ware ML, Flemington EK. 2016. A comprehensive next generation sequencing-based virome assessment in brain tissue suggests no major virus-tumor association. Acta Neuropathol Commun 4:71. doi: 10.1186/s40478-016-0338-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hu X, Yu J, Crosby SD, Storch GA. 2013. Gene expression profiles in febrile children with defined viral and bacterial infection. Proc Natl Acad Sci U S A 110:12792–12797. doi: 10.1073/pnas.1302968110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ohyashiki JH, Takaku T, Ojima T, Abe K, Yamamoto K, Zhang Y, Ohyashiki K. 2005. Transcriptional profiling of human herpesvirus type B (HHV-6B) in an adult T cell leukemia cell line as in vitro model for persistent infection. Biochem Biophys Res Commun 329:11–17. doi: 10.1016/j.bbrc.2005.01.090. [DOI] [PubMed] [Google Scholar]
- 28.Nastke M-D, Becerra A, Yin L, Dominguez-Amorocho O, Gibson L, Stern LJ, Calvo-Calle JM. 2012. Human CD4+ T cell response to human herpesvirus 6. J Virol 86:4776–4792. doi: 10.1128/JVI.06573-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ogata M, Satou T, Kawano R, Takakura S, Goto K, Ikewaki J, Kohno K, Ikebe T, Ando T, Miyazaki Y, Ohtsuka E, Saburi Y, Saikawa T, Kadota J. 2010. Correlations of HHV-6 viral load and plasma IL-6 concentration with HHV-6 encephalitis in allogeneic stem cell transplant recipients. Bone Marrow Transplant 45:129–136. doi: 10.1038/bmt.2009.116. [DOI] [PubMed] [Google Scholar]
- 30.Caserta MT, Hall CB, Schnabel K, Lofthus G, Marino A, Shelley L, Yoo C, Carnahan J, Anderson L, Wang H. 2010. Diagnostic assays for active infection with human herpesvirus 6 (HHV-6). J Clin Virol 48:55–57. doi: 10.1016/j.jcv.2010.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hill JA, Magaret AS, Hall-Sedlak R, Mikhaylova A, Huang M-L, Sandmaier BM, Hansen JA, Jerome KR, Zerr DM, Boeckh M. 2017. Outcomes of hematopoietic cell transplantation using donors or recipients with inherited chromosomally integrated HHV-6. Blood 130:1062–1069. doi: 10.1182/blood-2017-03-775759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zerr DM, Gupta D, Huang M-L, Carter R, Corey L. 2002. Effect of antivirals on human herpesvirus 6 replication in hematopoietic stem cell transplant recipients. Clin Infect Dis 34:309–317. doi: 10.1086/338044. [DOI] [PubMed] [Google Scholar]
- 33.Trapnell C, Pachter L, Salzberg SL. 2009. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Anders S, Pyl PT, Huber W. 2015. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leisenring WM, Martin PJ, Petersdorf EW, Regan AE, Aboulhosn N, Stern JM, Aker SN, Salazar RC, McDonald GB. 2006. An acute graft-versus-host disease activity index to predict survival after hematopoietic cell transplantation with myeloablative conditioning regimens. Blood 108:749–755. doi: 10.1182/blood-2006-01-0254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Øster B, Höllsberg P. 2002. Viral gene expression patterns in human herpesvirus 6B-infected T cells. J Virol 76:7578–7586. doi: 10.1128/JVI.76.15.7578-7586.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.