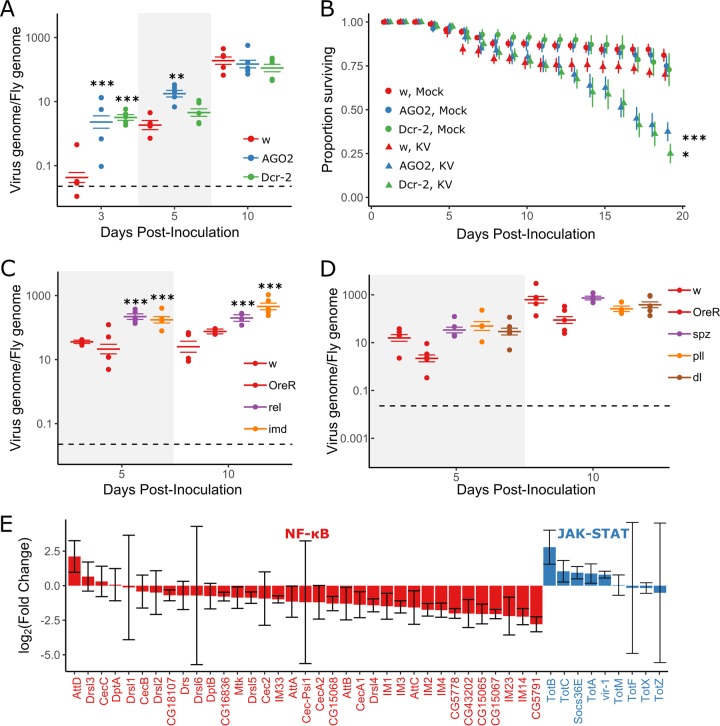
FIG 1.
RNAi and Imd pathways provide antiviral defense against Kallithea virus. Mutants for RNAi (A and B) and NF-κB (C and D) pathways were assayed for viral titer (A, C, and D) and mortality (B) following KV infection. Oregon R (OreR) and w1118 flies were used as wild-type controls. Viral titer was measured by qPCR, relative to Rpl32 DNA, where each data point represents a vial of 5 flies, and colored horizontal lines correspond to the mean titer and associated SE (A, C, and D). Horizontal dotted lines (A, C, and D) represent the amount of virus injected. (B) RNAi mutants (AGO2 and Dcr2) and w1118 controls were injected with chloroform-treated KV (mock) or KV, and survival was monitored each day. Each point is the mean number of surviving flies across 10 vials of 10 flies, with associated standard errors. (E) Log-transformed fold changes of presumed NF-κB-responsive genes (colored red; Cecropin, Diptericin, Attacin, Metchnikowin, Drosomycin and Drosomycin-like genes, Bomamins [i.e., IM1, CG18107, IM2, IM3, CG15065, CG15068, CG43202, CG16836, CG5778, IM23, CG15067, and CG5791)], and other IM genes) and JAK-STAT-responsive genes (colored blue; Socs36E, vir-1, and Turandot [Tot] family) following KV infection of OreR flies at 3 dpi, relative to uninfected controls (ERP023609; n = 5 libraries per treatment, with n = 10 flies per library [32]). Error bars show SEMs. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (statistical tests were performed in MCMCglmm).