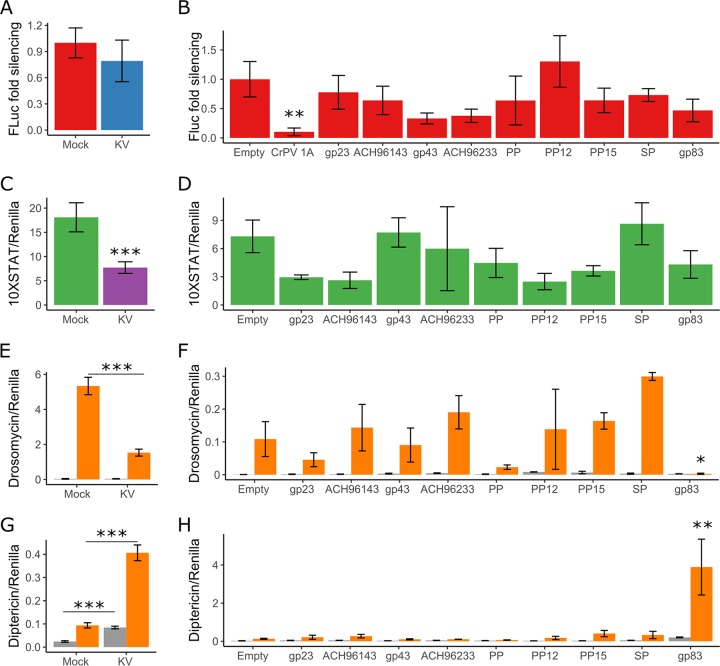
FIG 3.
Kallithea virus gp83 suppresses Toll and induces Imd signalling. The ability of KV (4 dpi) and 9 highly expressed KV genes to inhibit the RNAi (A and B), JAK-STAT (C and D), Toll (E and F), and Imd (G and H) pathways was assessed. For RNAi suppression assays (A and B), RNAi efficiency was assessed by transfecting S2 cells with plasmids expressing FLuc and, as a normalization control, Renilla luciferase (RLuc), along with dsRNA targeting either FLuc or GFP. Data are expressed as fold silencing in cells treated with GFP dsRNA relative to those treated with FLuc dsRNA, normalized to 1 in mock-infected cells. The CrPV suppressor of RNAi, protein 1A, was used as a positive control (data combined from 2 experiments). For JAK-STAT suppression assays (C and D), S2 cells were transfected with a plasmid encoding FLuc under the control of 10 STAT binding sites (10×STAT-FLuc). In contrast to the JAK-STAT pathway, the Toll and Imd pathways are not endogenously active in S2 cells (gray bars in E, F, G, and H) but can be activated by expression of TollLRR (orange bars in E and F) or PGRP-LC (orange bars in G and H). For Toll suppression assays (E and F), S2 cells were transfected with the Drs-FLuc reporter, encoding FLuc under the control of a Drosomycin promoter, with either pAc5.1-TollLRR or an empty control plasmid (gray bars). For Imd suppression assays (G and H), S2 cells were transfected with the Dpt-FLuc reporter, encoding FLuc under the control of a Diptericin promoter, with either pMT (empty) or pMT-PGRP-LC. All FLuc luciferase values were normalized to RLuc values, driven by a constitutively active Actin promoter from a cotransfected plasmid. PP, putative protein; SP, serine protease. Error bars show SEMs, calculated from 5 biological replicates for panels A, C, E, and G and at least 3 biological replicates for panels B, D, F, and H.