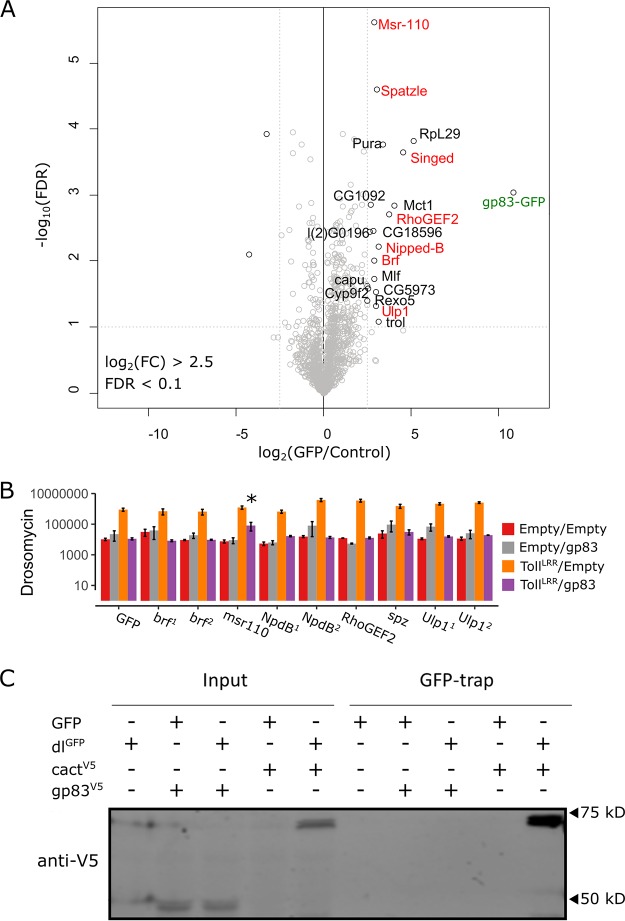
FIG 6.
Identification of host interactors of gp83. (A) Identification of gp83 interacting proteins in S2 cell lysates by label-free quantitative (LFQ) mass spectrometry. Permutation-based FDR-corrected t tests were used to determine proteins that are statistically enriched in gp83-GFP immunoprecipitation (IP). The log2 LFQ intensity of gp83-GFP IP over control IP (cells that do not express gp83-GFP) is plotted against the −log10 FDR. The gp83-GFP bait (labeled in green) and interactors with an enrichment of fold change of >2.5; −log10 FDR values of >1 are indicated. (B) Drs-FLuc expression was measured following cotransfection of pAc-gp83, pAc-TollLRR, or empty control plasmids, along with dsRNA targeting brf, msr-110, Nipped-B, RhoGEF2, spätzle, and Ulp1 (labeled red in panel A), with dsRNA targeting GFP as a control. Genes are superscripted with “1” or “2” when two independent dsRNAs were used to knock down the gene. Although msr-110 knockdown appears to partially rescue gp83 immunosuppression, subsequent experiments did not reproduce this effect. Error bars represent SEMs (n = 3). Statistical tests were performed in MCMCglmm. (C) V5-tagged gp83 or V5-tagged cact (an IκB protein known to interact with dl) were expressed alongside GFP-tagged dl or GFP and GFP-associated complexes were immunoprecipitated with GFP-trap magnetic beads and analyzed by Western blotting using V5 antibodies. Note that cact appears to be stabilized when coexpressed with dl compared to when it is expressed alone.