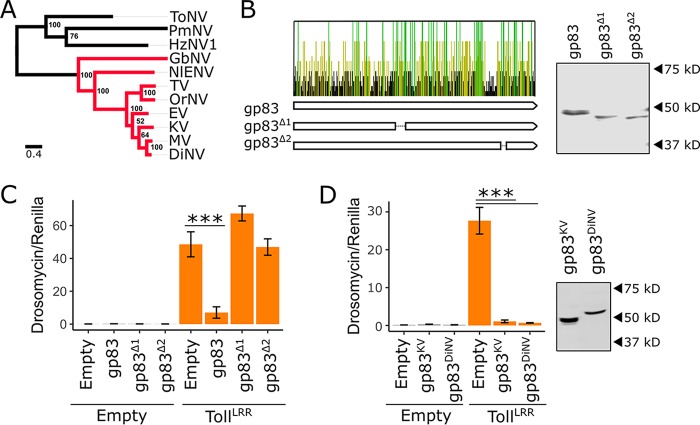
FIG 8.
The immunosuppressive function of gp83 is evolutionarily conserved. (A) Maximum likelihood phylogeny inferred from a protein alignment of nudivirus-encoded DNA polymerase B using PhyML (83), with an LG substitution model and gamma-distributed rate parameter. Support for each node was assessed by bootstrapping, and the scale bar represents substitutions per site. Nudivirus species that encode gp83 homologs are colored in red. (B) Conservation of the gp83 amino acid sequence across 7 species of nudivirus (all red-labeled viruses in panel A, except the endogenized virus NlENV). Each bar represents an amino acid, and bars are colored yellow if the residue is conserved in ≥50% of the species, green if conserved in 100% of the species, and black if conserved in <50% of the species. Two V5-tagged gp83 constructs were created with deletions that span regions with an excess of conserved residues: gp83Δ1 and gp83Δ2. Western blotting and subsequent V5 antibody staining show that both deletion constructs accumulate to levels similar to those of full-length gp83 following transfection of S2 cells. (C) Full-length gp83, gp83Δ1, or gp83Δ2 was coexpressed with TollLRR, and Drs-FLuc expression was measured relative to pAct-FLuc expression. (D) V5-tagged gp83 from KV and DiNV were coexpressed with TollLRR to assess suppression of Drs-FLuc expression (relative to pAct-FLuc expression) in D. melanogaster S2 cells. Western blot analysis using V5 antibody was used to confirm gp83 expression. Error bars show SEMs (n = 5 biological replicates).