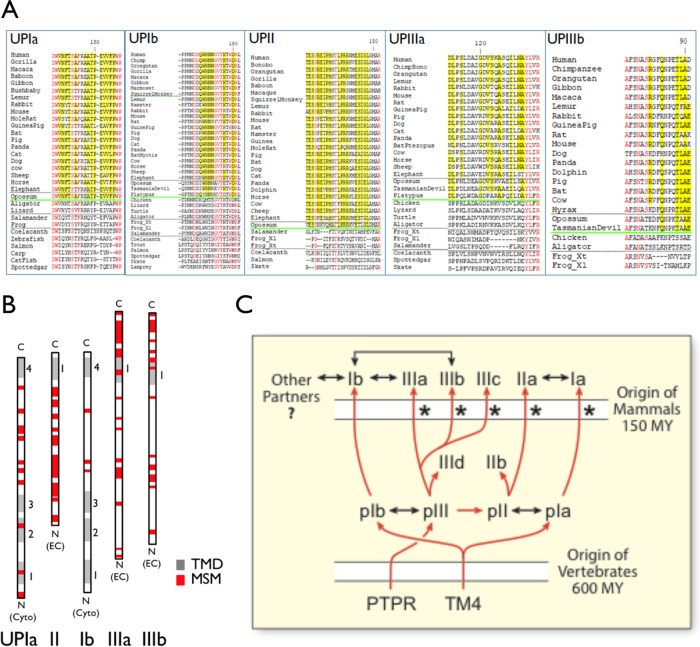
FIGURE 8:
Mammal-specific amino acid residues and motifs in uroplakins. (A) Examples of mammal-specific residues (yellow) and amino acid residues that are shared by ALL uroplakins (“all-uroplakin-residues or AUR; red) from UPIa, Ib, II, IIIa, and IIIb of ∼20 mammals (17 placental and three nonplacental) and 10 nonmammals (Desalle et al., 2014). Mammal-specific residues are defined as those that are present in >90% of mammals and <30% of nonmammals. The horizontal green line demarcates the sequences of the mammals (above) and nonmammal vertebrates. (B) The overall location of mammal-specific motifs (MSM; red box; greater than three consecutive residues, five with a single interruption, or six with two interruptions) in uroplakins as indicated. TMD (transmembrane domain). (C) Model depicting the evolution of all the known uroplakin genes. Symbols: red arrows (genealogical relationship), two-headed black arrows (protein–protein interaction), asterisk (a strong pattern of significant skew towards dN/dS>1.0 suggesting possible selection that accompanies the duplication events that produced the paralogue group) (Desalle et al., 2014), phosphotyrosine phosphatase receptor (PTPR) (Chicote et al., 2017), tetraspanin precursor (TM4), million years (MY). See main text for details.