Abstract
Microtubule nucleation was uncovered as a key principle of spindle assembly. However, the mechanistic details about microtubule nucleation and the organization of spindle formation and symmetry are currently being revealed. Here we describe the function of coiled-coil domain containing 61 (Ccdc61), a so far uncharacterized centrosomal protein, in spindle assembly and symmetry. Our data describe that Ccdc61 is required for spindle assembly and precise chromosome alignments in mitosis. Microtubule tip-tracking experiments in the absence of Ccdc61 reveal a clear loss of the intrinsic symmetry of microtubule tracks within the spindle. Furthermore, we show that Ccdc61 controls the centrosomal localization of centrosomal protein of 170 kDa (Cep170), a protein that was shown previously to localize to centrosomes as well as spindle microtubules and promotes microtubule organization and microtubule assembly. Interestingly, selective disruption of Ccdc61 impairs the binding between Cep170 and TANK binding kinase 1, an interaction that is required for microtubule stability. In summary, we have discovered Ccdc61 as a centrosomal protein with an important function in mitotic microtubule organization.
INTRODUCTION
The assembly of a bipolar spindle is essential for the accurate segregation of chromosomes during mitosis and meiosis and relies on the tightly regulated nucleation of microtubules (MTs). Formation of bipolar mitotic spindles with bioriented chromosomes ensures the faithful segregation of a complete set of chromosomes to each daughter cell. Proper attachment of MTs and kinetochores are monitored by the spindle assembly checkpoint (SAC). If all kinetochores have established a proper and stable attachment to the spindle, the SAC is satisfied and thus silenced ensuring proper mitotic progression (reviewed in Prosser and Pelletier [2017]).
The main MT-organizing center in mammalian cells is the centrosome. It inherits MT nucleation capacity and influences thereby MT-dependent processes such as transport of organelles, cell motility, cell polarity, cell division, and ciliogenesis (Conduit et al., 2015). A functional centrosome contains two MT-based barrel-shaped centrioles, which are surrounded by a well-organized protein matrix called the pericentriolar material (PCM). The two centrioles present differ from one another in both structure and age. The older one of the two centrioles, which is known as the mother centriole, harbors distal and subdistal appendages, structures that are important for MT anchorage (Chretien et al., 1997; Bornens, 2002; Meraldi and Nigg, 2002). In contrast, the younger centriole (daughter centriole) lacks distal and subdistal appendages. MTs nucleate from the PCM, even in the absence of appendages, and are anchored at the distal end of the mother centriole (Piel et al., 2000). The amount of PCM material correlates with the MT nucleation capacity of the centrosome. Since the PCM extends as cells prepare to enter mitosis, a process known as centrosome maturation, MT nucleation capacity increases positively influencing mitotic spindle assembly dynamics (Conduit et al., 2015).
Centriolar satellites were identified as electron-dense structures in the vicinity of centrosomes (Kubo et al., 1999). They contain proteins involved in the recruitment of core centriolar factors and have been implicated in centrosome maturation, duplication, ciliogenesis as well as MT organization and mitotic spindle formation (Tollenaere et al., 2015). Centriolar satellites require an intact MT-network and a direct interaction with dynein/dynactin motor proteins for correct localization and cargo delivery (Barenz et al., 2011; Tollenaere et al., 2015). Pericentriolar material 1 protein (PCM1) was the first identified centriolar satellite component and acts a scaffold for centrosomal proteins for interphase MT organization (Balczon et al., 1994; Balczon et al., 2002; Dammermann and Merdes, 2002; Kubo and Tsukita, 2003).
The mother centriole-localized centrosomal protein of 170 kDa (Cep170) is associated with subdistal appendages and the proximal ends of centrioles in interphase (Guarguaglini et al., 2005; Mazo et al., 2016). In mitosis, Cep170 localizes to the spindle apparatus and is heavily phosphorylated by Plk1 (Guarguaglini et al., 2005). Depletion of Cep170 affects MT cytoskeleton organization and cell shape in interphase and seems thereby directly involved in the regulation of MT organization, assembly and stability (Guarguaglini et al., 2005; Welburn and Cheeseman, 2012; Dumoux et al., 2015). Additionally, Cep170 gets phosphorylated by and forms a complex with TANK binding kinase 1 (TBK1) (Pillai et al., 2015). TBK1 is necessary for Cep170 centrosomal localization and disruption of TBK1/Cep170 binding increases MT stability resulting in mitotic defects (Pillai et al., 2015).
In the presented study, we report the characterization of coiled-coil domain containing 61 (Ccdc61), a protein of around 70 kDa. We show that Ccdc61 is a protein localizing at the centrosome. Ccdc61 is involved in mitotic spindle assembly and chromatin alignment by regulating the organization of spindle MTs into a symmetrical structure. Furthermore, we show that Ccdc61 depletion affects Cep170 maintenance on interphase and mitotic centrosomes; suggesting a function of Ccdc61 as a centrosomal anchorage factor for Cep170 and thereby influencing MT organization.
RESULTS
Ccdc61 is a centrosomal protein with a satellite-like staining pattern
Ccdc61 was identified in proteomic approaches to define the constituents of human centrosomes (Jakobsen et al., 2011; Gupta et al., 2015; Bauer et al., 2016) implicating a so far unknown function of Ccdc61 at centrosomes. We initially analyzed the localization of overexpressed green fluorescent protein (GFP)- or FLAG-tagged Ccdc61 in RPE1 cells (Supplemental Figure S1A) and further studied the localization of the GFP-tagged mouse homolog in a stable HeLa cell line in immunofluorescence (IF) (Supplemental Figure S1B). Costaining with γ-tubulin, pericentrin, and PCM1 revealed a centrosomal localization and a granular-like staining in close proximity to centrosomes (Supplemental Figure S1, A and B). The granular localization pattern of Ccdc61 resembles the subcellular localization described for centriolar satellite components (Barenz et al., 2011; Tollenaere et al., 2015).
To further explore the localization of endogenous Ccdc61, we generated and characterized a monoclonal antibody against full-length human Ccdc61 (Supplemental Figure S1, C and D). Endogenous Ccdc61 localization in interphase cells reveals, similar as the overexpressed protein, a granular-like staining at centrosomes and in their close proximity (Figure 1A and Supplemental Figures S1C and S2A). To further confirm the specificity of the Ccdc61 antibody generated, we performed small interfering RNA (siRNA)-mediated down-regulation experiments of Ccdc61, that resulted in a loss of the granular and centrosomal staining in IF (Supplemental Figure S1C). As we failed to detect endogenous Ccdc61 in Western blots using our antibody, we have quantified the amount of endogenous down-regulation in IF (Supplemental Figure S1C). Additionally, we were able to enrich endogenous Ccdc61 after immunoprecipitation from cell lysates and also detect Ccdc61 overexpression with our antibody supporting the specificity (Supplemental Figure S1D).
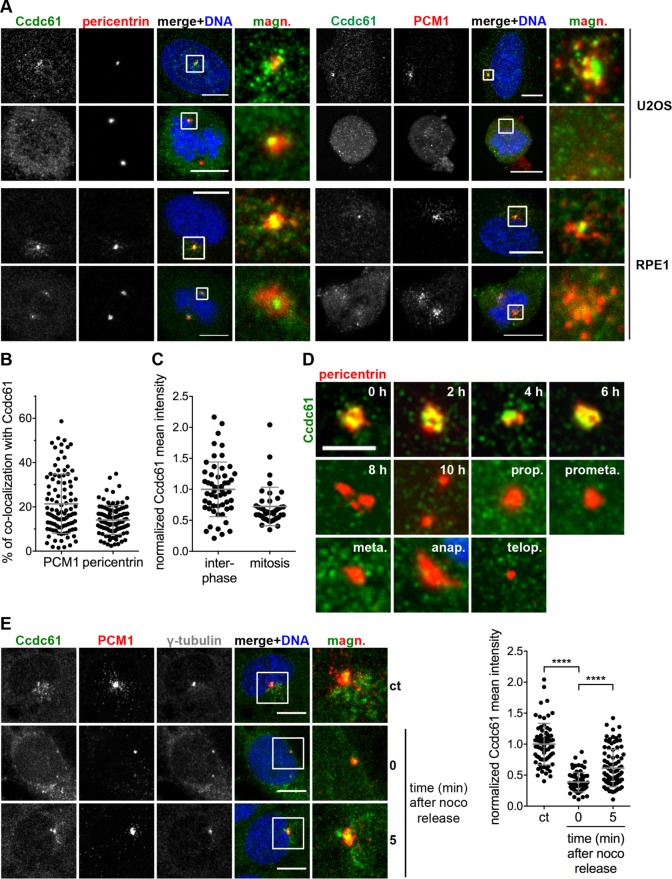
FIGURE 1:
Ccdc61 localizes MT-dependent at centrosomes and in their close proximity. (A) Costaining of endogenous Ccdc61 (green) with pericentrin or PCM1 (red) in U2OS and RPE1 cells. Chromatin is visualized by Hoechst33258 staining (blue). Magnified panels (magn.) show enlarged views of the boxed regions. Bar, 10 µm. (B) Graph shows the percentage of colocalization of endogenous Ccdc61 with either PCM1 or pericentrin in U2OS cells. Therefore, signals of PCM1 or pericentrin were used as a mask to measure the corresponding Ccdc61 signal intensity. n = 100 cells for each colocalization. Data represent mean value ± SD. (C) Endogenous Ccdc61 signal was quantified in a 3-µm2 circle around the centrosome in interphase or mitotic U2OS cells. Interphase Ccdc61 signal was normalized to 1.0. Interphase n = 54, mitosis = 40 centrosomes. Data represent mean value ± SD. (D) HeLa cells were released from a thymidine block and samples taken at indicated time points to localize endogenous Ccdc61 (green) at centrosomes by costaining pericentrin (red). Bar, 4 µm. (E) RPE1 cells were subjected to nocodazole (noco) to depolymerize MTs. After nocodazole washout, cells were fixed at the indicated time points and stained with antibodies directed against Ccdc61 (green), PCM1 (red), and γ-tubulin (gray). Magnified panels (magn.) show enlarged views of the boxed areas. Bar, 10 µm. Quantification shows normalized mean intensity of Ccdc61 in a 3-µm2 circle around the centrosome. The intensity was normalized to the nocodazole-untreated sample (ct: control). Data represent mean ± SD. from three independent experiments, n > 69 cells. ****p < 0.0001 (unpaired Student’s t test).
In interphase U2OS, RPE1, and HeLa cells endogenous Ccdc61 partially colocalizes with pericentrin and the centrosomal satellite component PCM1 (Figure 1, A and B, and Supplemental Figure S2A). However, endogenous Ccdc61 seems to be significantly released into the cytoplasm during mitosis, since Ccdc61 is almost completely lost from its centrosomal/scattered localization in mitotic U2OS, RPE1 and HeLa cells (Figure 1, A and C, and Supplemental Figure S2A). This observation shows strong similarities with previous centriolar satellite studies, showing that these structures gradually dissolve when cells enter mitosis (Kubo and Tsukita, 2003). The gradual loss of endogenous Ccdc61 from the vicinity of centrioles during mitosis was further analyzed in thymidine blocked and released HeLa cells (Figure 1D). Endogenous Ccdc61 is dispersed from these centrosomal/satellite-like structures with the onset of centrosome separation in early G2 and is almost completely released into the cytoplasm or degraded during mitosis (Figure 1D). In contrast to the endogenous protein, overexpressed Ccdc61 maintains its localization in mitosis in close proximity to the centrosome (Supplemental Figure S1B).
To test whether Ccdc61 requires an intact MT network for correct localization, we depolymerized MTs in RPE1 and U2OS cells by nocodazole-treatment and monitored the localization pattern of Ccdc61 (Figure 1E and Supplemental Figure S2B). Interestingly, Ccdc61 interphase localization changes significantly after MT depolymerization (Figure 1E and Supplemental Figure S2B), whereby Ccdc61 granules loose their close localization to the centrosomes. However, 5 min after MT regrowth, the Ccdc61 signal increased again at the centrosomes (Figure 1E). This MT-dependent localization pattern observed for Ccdc61 has previously been characterized for centriolar satellite components (Tollenaere et al., 2015).
Both the structure of centriolar satellites and the localization of satellite proteins depends on the presence of the core satellite protein PCM1 that forms a structural platform for satellites (Stowe et al., 2012). Since Ccdc61 exhibit significant similarities with satellite proteins regarding its localization pattern and MT dependency, we analyzed whether Ccdc61 localization also depends on the presence of PCM1. To our surprise, the localization of Ccdc61 did not change on PCM1 depletion by RNA-interference suggesting that the satellite-like staining pattern of Ccdc61 was not dependent on PCM1 (Supplemental Figure S2C). Therefore, Ccdc61, despite its scattered localization around centrosomes, does not seem to be a bona fide centriolar satellite component. As this was an interesting observation, we decided to further investigate its direct function.
Ccdc61 promotes correct chromatin alignment and spindle assembly
Next, we addressed the possible impact of Ccdc61 on the formation of the mitotic spindle. Therefore, we performed live cell imaging with U2OS cells stably expressing fluorescently labeled histone 2B (H2B) and α-tubulin that synchronously proceeded through mitosis. The cells were treated with control (ct) or Ccdc61 siRNA Oligo2 (O2), or with a combination of Ccdc61 siRNA Oligo2 and the siRNA-resistant Ccdc61 construct (O2R). We first analyzed the timing of cells going through mitosis, starting from nuclear envelope breakdown (judged by the state of chromosome condensation) until reaching telophase (Figure 2A). Data analysis identified a significant delay in mitotic progression after loss of Ccdc61, that could also been rescued in the presence of the siRNA-resistant construct (Figure 2A, Supplemental Figure S3A, and Supplemental Movies 1–6). The overall mitotic duration increased significantly from approximately 58 min in control-treated cells to 77 min in Ccdc61 depleted cells (Figure 2A). A similar increase in mitotic timing was observed in HeLa mCherry-H2B α-tubulin–monomeric enhanced GFP (mEGFP) cells, treated with control, Oligo1, or Oligo2 siRNA (Supplemental Figure S3B). Further quantification of mitosis revealed that the increased mitotic duration was caused by a significant prolonged metaphase-to-anaphase transition (from approx. 26 min in control siRNA-treated cells to 39 min in Ccdc61-depeted cells; Figure 2A), probably induced by an activated SAC, as shown by Mad2 staining (Supplemental Figure S3C).
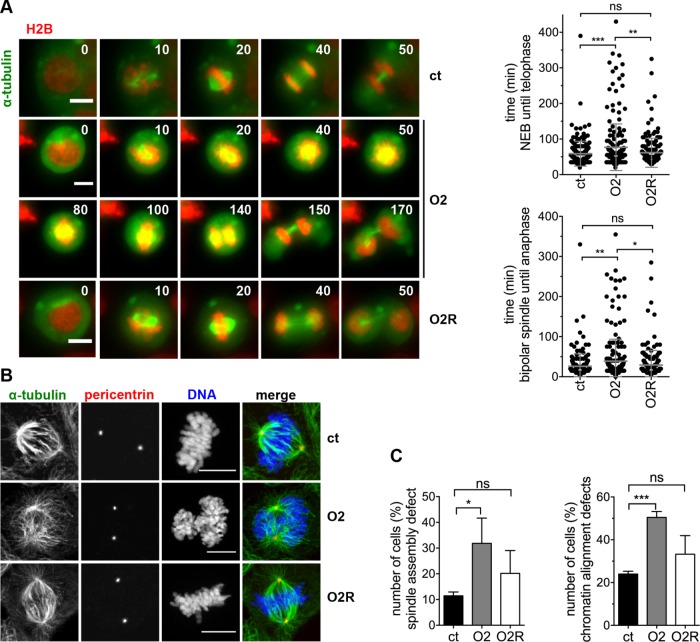
FIGURE 2:
Ccdc61 is required for proper timing of mitosis, chromatin alignment and spindle asembly. (A) U2OS cells stably expressing tagged α-tubulin (green) and H2B (red) were imaged every 5 min for 15–19 h 72 h after siRNA transfection (ct: control; O2: Oligo2; O2R: Oligo2 + FLAG-Ccdc61 siRNA-resistant). Representative images from the movies were cropped out from the original time series (time in min). Bar, 10 µm. Quantification of the indicated treatments show the mitotic duration from nuclear envelope breakdown (start of chromosome condensation and movement of centrosomes) until telophase (decondensation of chromosomes) (top panel) and duration from bipolar spindle formation to anaphase (bottom panel). Data represent mean ± SD from four independent experiments, n > 167 cells. ns: not significant, *p < 0.05, **p < 0.01, ***p < 0.001 (unpaired Student’s t test). (B) IF analysis of mitotic figures in U2OS cells indicated treatments. Mitotic spindles and centrosomes are visualized by antibody staining directed against α-tubulin (green) and pericentrin (red), costained with Hoechst33258 (DNA; blue). Bar, 10 µm. (C) U2OS cells stably expressing tagged α-tubulin and H2B were imaged every 5 min for 15–19 h 72 h after indicated siRNA transfection. Quantification shows the analyzed spindle assembly defects and chromatin alignment defect. Spindle assembly defects include various problems of the cell to assemble a properly formed spindle (including, e.g., multipolar spindle but also bipolar spindles struggling to align DNA as indicated in Figure 2, A and B, for O2 treatment). Chromatin alignment defects include disturbed formation of a properly arranged metaphase plate, e.g., lagging chromosomes. Data represent mean ± SD from three independent experiments, n > 76 cells. ns: not significant, *p < 0.05, ***p < 0.001 (paired Student’s t test).
Movie S1.
Time lapse imaging of 2 control (ct) siRNA treated U2OS H2B-GFP α-tubulin-mCherry cells. Imaging was performed every 5 min on an inverted microscope (Observer.Z1; Zeiss) using a 20× objective lens. Time is indicated in h:min. Bar, 10 μm.
Movie S2.
Time lapse imaging of 2 control (ct) siRNA treated U2OS H2B-GFP α-tubulin-mCherry cells. Imaging was performed every 5 min on an inverted microscope (Observer.Z1; Zeiss) using a 20× objective lens. Time is indicated in h:min. Bar, 10 μm.
Movie S3.
Time lapse imaging of 2 Ccdc61 Oligo2 siRNA treated U2OS H2B-GFP α-tubulinmCherry cells. Imaging was performed every 5 min on an inverted microscope (Observer.Z1; Zeiss) using a 20× objective lens. Time is indicated in h:min. Bar, 10 μm.
Movie S4.
Time lapse imaging of 2 Ccdc61 Oligo2 siRNA treated U2OS H2B-GFP α-tubulinmCherry cells. Imaging was performed every 5 min on an inverted microscope (Observer.Z1; Zeiss) using a 20× objective lens. Time is indicated in h:min. Bar, 10 μm.
Movie S5.
Time lapse imaging of 2 U2OS H2B-GFP α-tubulin-mCherry cells treated with Ccdc61 Oligo2 siRNA and FLAG-Ccdc61 siRNA-resistant construct. Imaging was performed every 5 min on an inverted microscope (Observer.Z1; Zeiss) using a 20× objective lens. Time is indicated in h:min. Bar, 10 μm.
Movie S6.
Time lapse imaging of 2 U2OS H2B-GFP α-tubulin-mCherry cells treated with Ccdc61 Oligo2 siRNA and FLAG-Ccdc61 siRNA-resistant construct. Imaging was performed every 5 min on an inverted microscope (Observer.Z1; Zeiss) using a 20× objective lens. Time is indicated in h:min. Bar, 10 μm.
To learn whether spindle formation failure may cause the delay in mitosis, we performed IF analysis of the mitotic spindle via staining with α-tubulin and pericentrin and observed, that the bipolarity of the formed spindle was not impaired (Figure 2B). However, the cells seem to have difficulties to establish a proper spindle-chromosome connection. Additionally, staining with CREST to visualize kinetochores and α-tubulin for MTs indicated that fewer MTs are properly attached to kinetochores upon depletion of Ccdc61 (Supplemental Figure S3D). To further analyze this hypothesis, we investigated spindle formation by analyzing spindle assembly and chromatin alignment in live cell imaging of H2B- and α-tubulin labeled U2OS cells. Quantifications revealed in fact a significant defect of mitotic spindle assembly and chromatin alignment after loss of Ccdc61 (Figure 2C). To confirm the specificity of the Ccdc61 depletion, rescue experiments were performed by overexpression of a siRNA-resistant version of Ccdc61 that successfully reverted the observed spindle assembly and chromatin alignment defects (Figure 2, B and C, and Supplemental Figure S3, C and D). Collectively, these results indicate that Ccdc61 influences the correct organization of spindle MTs and chromatin alignment affecting mitotic progression and correct chromosome segregation.
Ccdc61 promotes MT nucleation and the intrinsic symmetry of MTs in mitotic spindles
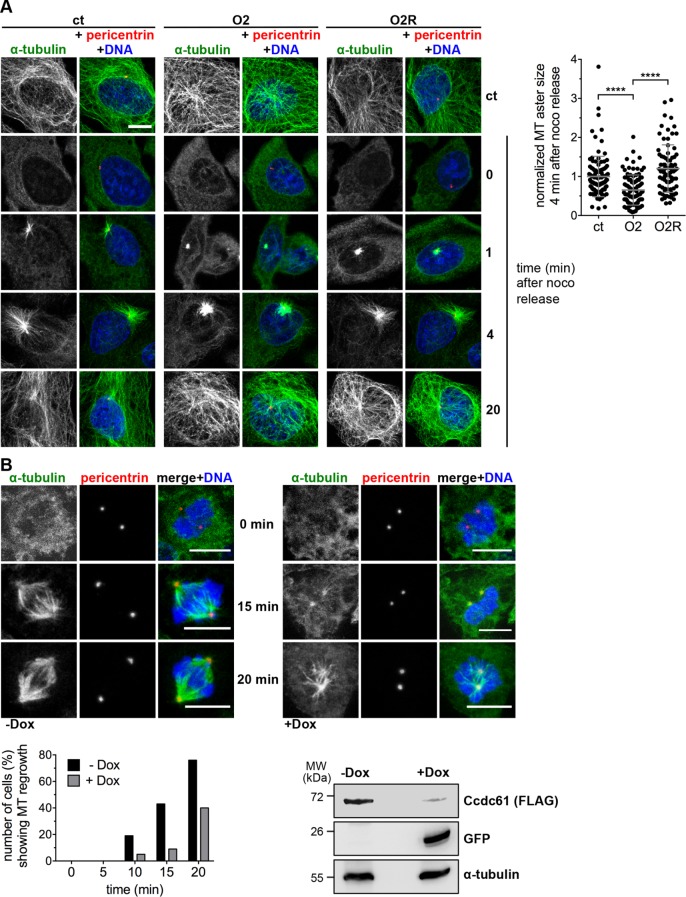
To evaluate the function of Ccdc61 in spindle formation and MT organization in more detail, we performed MT regrowth assays in U2OS cells that were exposed to nocodazole and then released from the block (Figure 3A) in cells treated with ct, O2 or O2R. We analyzed MT aster formation at an early time point and found that MTs were detectable 1 min after release in control cells but greatly reduced in Ccdc61 siRNA-treated cells (Figure 3A). After 4 min of MT regrowth, MT asters were fully assembled in control cells whereas the size of MT aster showed a clear reduction after loss of Ccdc61. Expressing a siRNA-resistant construct of Ccdc61 could rescue the loss of Ccdc61 (Figure 3A). Additionally, we studied MT organization in mitotic HeLa cells. For this, we used a miRNA tet-on HeLa cell line expressing Ccdc61, Oligo2, and GFP as a control on doxycycline induction (Figure 3B). Although MTs regrew in control cells and formed a spindle within 10 min, only inefficient MT growth and spindle formation was observed in Ccdc61 depleted cells (Figure 3B). Together, these data indicate that Ccdc61 is rate-limiting for efficient MT nucleation and organization in mitosis; explaining the initially observed chromatin alignment and mitotic spindle assembly defects.
FIGURE 3:
Ccdc61 promotes MT nucleation. (A) U2OS cells were treated as indicated and subjected to nocodazole (noco) for 2 h to depolymerize MTs. After nocodazole washout, cells were harvested at the indicated time points and stained with antibodies directed against α-tubulin (green) and pericentrin (red). DNA was costained with Hoechst33258 (blue). Cells without nocodazole exposure were used as a control (ct). Bar, 10 µm. Quantification indicates the MT aster size formation 4 min after nocodazole release compared with the control (without nocodazole treatment) (normalized to 1.0). Data represent mean ± SD from three independent experiments, n > 80 cells. ****p < 0.0001 (unpaired Student’s t test). (B) HeLa S/A Ccdc61 miRNA Oligo2 tet on cells were used to perform a MT regrowth assay. α-tubulin was used to visualize mitotic spindles (green), pericentrin (red) used to stain the centrosomes, and Hoechst33258 to stain DNA (blue). Ccdc61 miRNA O2 expression was induced by the addition of 1 µg/ml doxycycline for 72 h (+Dox) prior cold treatment. For MT depolymerization cells with or without Dox addition were incubated at 4°C for 45 min, and then reincubated at 37°C for the indicated time points. Bar, 10 µm. Twenty mitotic cells from each situation (–Dox vs. +Dox) were visually inspected for correct spindle reassembly and MT regrowth at the indicated time points. Western blot shows the efficiency of indicated Ccdc61 miRNA that was tested by its ability to deplete transiently transfected FLAG-Ccdc61. α-tubulin served as a loading control and GFP as expression control of the miRNA construct.
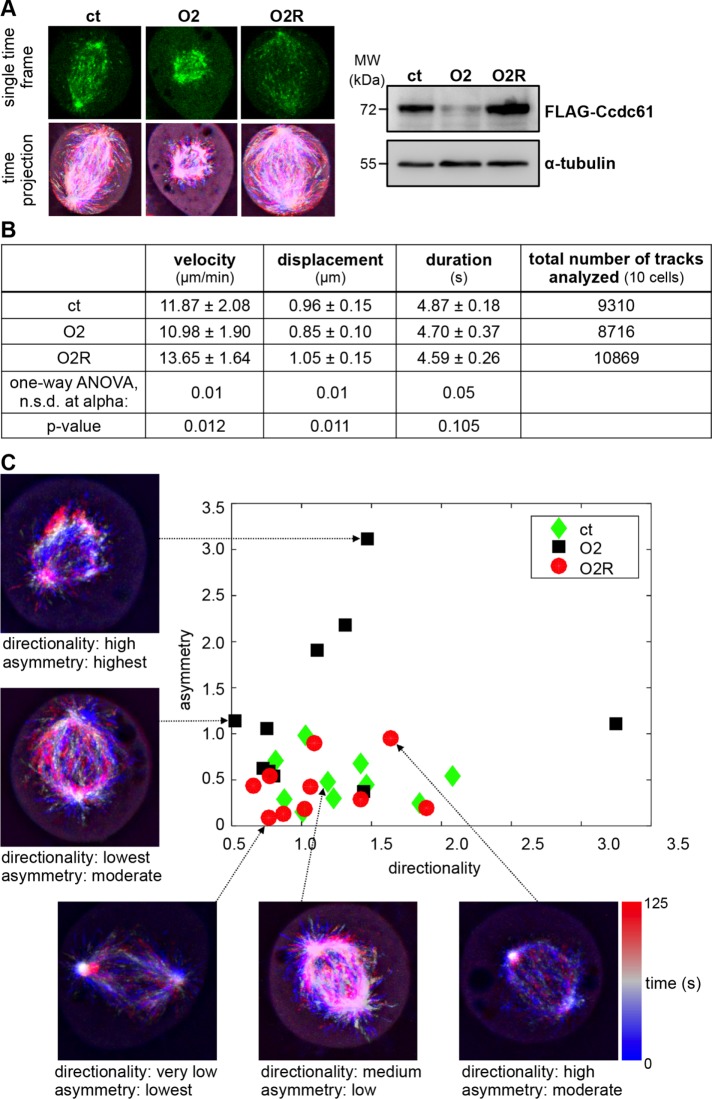
To quantitatively analyze the role of Ccdc61 in MT behavior and dynamics in mitosis, we imaged HeLa cells expressing EGFP–end-binding protein 3 (EB3) over time after the different siRNA treatments (ct, O2, O2R) and performed automated image analysis of MT plus ends (Applegate et al., 2011). Compared to previous studies (Sironi et al., 2011; Chinen et al., 2015), we acquired image series at faster frame rates (0.13 s) over a longer period of time in order to ensure reliable tracking information for a larger time contingent (Jafarpour and Lorenz, 2017). Interestingly, time projections of image series (Figure 4A) demonstrate variations in the symmetry and the directionality of MT growth paths in the differently treated cells. However, we found that key parameters of MT dynamics, including track velocity, duration, and displacement, were only slightly different after Ccdc61 down-regulation (Figure 4B).
FIGURE 4:
MT EB3-tracking measurements reveal a significant decrease in spindle symmetry in the absence of Ccdc61. (A–C) HeLa cells stably expressing EGFP-EB3 were used for EB3-tracking experiments. Cells were either treated with control (ct), Ccdc61 O2 siRNA (O2), or a combination of Ccdc61 O2 siRNA together with FLAG-Ccdc61 siRNA-resistant construct (O2R). To study mitotic properties, like directionality and orientation, the individual EB3 tips were imaged every 0.13 s and analyzed via an automated process. (A) Example images of a single time frame (top row) and a time projection from 125 s recordings (bottom row) of HeLa cells treated as indicated. Western blot shows the efficiency of indicated Ccdc61 Oligo that was tested by its ability to deplete transiently transfected FLAG-Ccdc61 or siRNA-resistant FLAG-Ccdc61 (O2R). α-tubulin served as a loading control. (B) EGFP-EB3 tracks from HeLa cells were analyzed as described in the Materials and Methods section and as published in the preprint (Jafarpour and Lorenz, 2017). The table represents velocity (µm/min), displacement (µm), duration (s) (mean ± SD.) and the total number of analyzed tracks (of 10 independent cells). The significance was calculated for the three conditions by a one-way ANOVA, and the alpha value at which there was no significant difference (n.s.d.) within the group as well as the p value is provided. (C) Shown are the results of calculated asymmetry and directionality indices of the indicated treated HeLa cells with 10 image series per group based on the histogram of calculated orientations (Supplemental Figure S4, B and C). Each mitosis event is characterized with two figures of merit, resulting in a single point. The y-axis represents the degree of asymmetry, whereby a low value indicates a symmetric direction of MT growth (low asymmetry) and a high value indicates an asymmetric direction of MT growth (reduced symmetry). The horizontal axis represents the directionality of MT growth, indicating different levels of point symmetry in MT growth paths.
Since Ccdc61 appeared to be important for efficient MT nucleation and organization (Figures 2 and 3), we decided to further explore MT behavior. To quantify the MT growth with comparable and conclusive indices, we developed a novel software tool, which enabled us to automatically measure geometrical MT features within spindles. By identifying preferred directions in MT growth among all tracks of a spindle, the software reduced each time-lapse image series to two indices of “directionality” and “asymmetry” (Figure 4C, Supplemental Figure S4, A–C, and Materials and Methods). Both indices function as descriptors of MT morphology. Exemplified by a normal bipolar spindle, the directionality is maximized (ordered) in meta-/anaphase, since the route of growing MTs is narrowed toward chromosomes. At the same time, the asymmetry index is minimized reflecting maximum MT symmetry. In earlier (prophase and prometaphase) or later (telophase) phases of mitosis, the growth of MT is less ordered (i.e., directional), and thus the directionality index is reduced (Figure 4C).
Analyzing the imaging data with the new software revealed that Ccdc61 directly influences the intrinsic symmetry of the MT tracks within the spindle, independent of track size and position (Figure 4, A and C). The asymmetry index was highest for cells depleted of Ccdc61 compared with the control. Furthermore, the significant decrease in spindle symmetry was rescuable by overexpression of a siRNA-resistant Ccdc61 construct, supporting the specificity of these findings (Figure 4C). Loss of spindle symmetry through less ordered growing MTs might affect the correct attachment of spindle MTs to chromatin in metaphase plates. Taken together these data indicate that the disorganization of spindle MTs cause the observed defects in spindle assembly and chromatin alignment (Figure 2) upon cellular depletion of Ccdc61.
Ccdc61 regulates the centrosomal anchorage of the MT binding protein Cep170
The depletion of single centriolar satellite components has very different consequences on the correct function of the satellite complex itself (Klinger et al., 2014). It affects, however, the correct localization or anchorage of single centrosomal factors, thereby affecting maturation, MT anchorage or mitotic spindle assembly. Since Ccdc61 may have similar functions as centriolar satellite proteins, we tested whether the observed defects for Ccdc61 depletion are a consequence of the mislocalization of a distinct subset of centrosomal or centriolar satellite components. To identify possible candidates in close vicinity to Ccdc61 or interacting with Ccdc61, we applied the proximity-dependent biotin identification (BioID) method followed by mass spectrometry (Supplemental Figure S5) (Roux et al., 2012). Because BioID does not require preservation of endogenous protein–protein interactions for their identification, cell lysis can be performed under denaturing conditions, allowing the detection of poorly soluble candidates, such as centrosome-associated proteins (Roux et al., 2012; Firat-Karalar and Stearns, 2015; Yeh et al., 2015). FLAG-Ccdc61-BirA* expression was induced with the addition of doxycycline in HEK293T cells and localized, as expected, in the vicinity of the centrosome (Supplemental Figure S5A). FLAG-Ccdc61-BirA* was expressed in the absence of biotin as a control (ct) and in the presence of biotin in G1/S (thymidine addition) or mitosis (nocodazole addition) arrested cells (Supplemental Figure S5B). We identified centrosomal and MT binding proteins, which associate with the mitotic spindle, including the centriolar satellite components PCM1 and Cep290 and the centriolar protein Cep170 (Supplemental Figure S5, C and D).
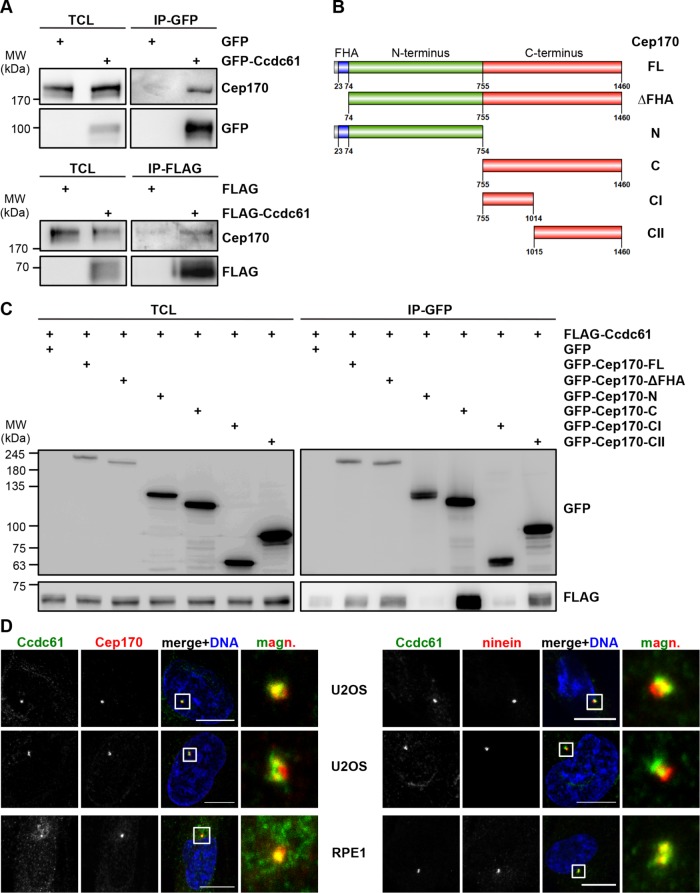
Interestingly, Cep170 was previously shown to regulate MT anchoring to the centrosome and thus directly influences MT assembly, organization and stability (Guarguaglini et al., 2005; Welburn and Cheeseman, 2012; Dumoux et al., 2015). To further validate the interaction of Ccdc61 with Cep170, we expressed GFP- or FLAG-tagged Ccdc61 in HEK293T cells followed by immunoprecipitation with FLAG or GFP antibodies. In both cases, we found endogenous Cep170 coprecipitating with Ccdc61 (Figure 5A). To map the interaction between the two proteins in more detail, we used truncated Cep170 fragments (Figure 5B) (Guarguaglini et al., 2005). Coexpression of full-length FLAG-Ccdc61 with GFP-Cep170 fragments in HEK293T cells followed by immunoprecipitation using antibodies against the GFP-tag showed that Ccdc61 interacted strongest with the Cep170 CII-terminal fragment 1015–1460 (Figure 5C). Interestingly, this fragment harbors both centrosomal and MT binding sites of Cep170 (Guarguaglini et al., 2005). We additionally observed a colocalization of Ccdc61 and Cep170 at centrosomes in interphase U2OS and RPE1 cells (Figure 5D, left panel).
FIGURE 5:
Ccdc61 binds to and colocalizes with Cep170. (A) Immunoprecipitation (IP) experiments using HEK293T cells expressing GFP-Ccdc61 or FLAG-Ccdc61 as bait. Western blot analysis shows total cell extract (TCL) as input control and corresponding GFP- and FLAG-IP fraction. GFP or FLAG antibody was used to visualize the overexpressed Ccdc61 and Cep170 antibody (mouse and rabbit) was used for detection of endogenous Cep170. (B) Schematic representation of Cep170 full length (FL) and Cep170 fragments: Cep170 missing the FHA domain (ΔFHA), Cep170 N-terminus (N), Cep170 C-terminus, Cep170 first half of C-terminus (CI) and Cep170 second half of C-terminus (CII) (Guarguaglini et al., 2005). (C) GFP-IP using HEK293T cells expressing GFP-Cep170 fragments as bait and FLAG-Ccdc61. Western blot shows TCL as input control and GFP-IP fraction. GFP antibody was used to visualize overexpressed Cep170 and FLAG antibody to visualize overexpressed Ccdc61. (D) Costaining of Ccdc61 (green) together with either (left) Cep170 or (right) ninein (both in red) antibodies in interphase U2OS and RPE1 cells. Chromatin was visualized by costaining with Hoechst33258 (blue). Magnified panels (magn.) show enlarged views of the boxed areas. Bar, 10 µm.
Cep170 localizes to subdistal appendages (Guarguaglini et al., 2005) and to the proximal ends of both centrioles, similar as it was shown for ninein (Bouckson-Castaing et al., 1996; Mazo et al., 2016). As Ccdc61 colocalizes with Cep170 (Figure 5D, left panel), we analyzed in more detail the localization of Ccdc61 at the centrioles. We performed costaining with the subdistal appendage compounds ninein, ODF-2 and the proximal centriolar protein C-Nap1. While ninein is localized to the subdistal appendages of mother centrioles and proximal ends of both centrioles, ODF-2 was reported to be localized at both distal and subdistal appendages (Nakagawa et al., 2001; Ishikawa et al., 2005). We observed only a weak colocalization with ODF-2 (Supplemental Figure S6A) and a more clear colocalization of Ccdc61 with ninein (right panel in Figure 5D) and the proximal centriolar marker C-Nap1 (Supplemental Figure S6B). From these data, we anticipate that Ccdc61 might also localize to the outer ring of subdistal appendages and the proximal end of centrioles, thereby resembling the localization pattern of Cep170 and ninein (Ishikawa et al., 2005).
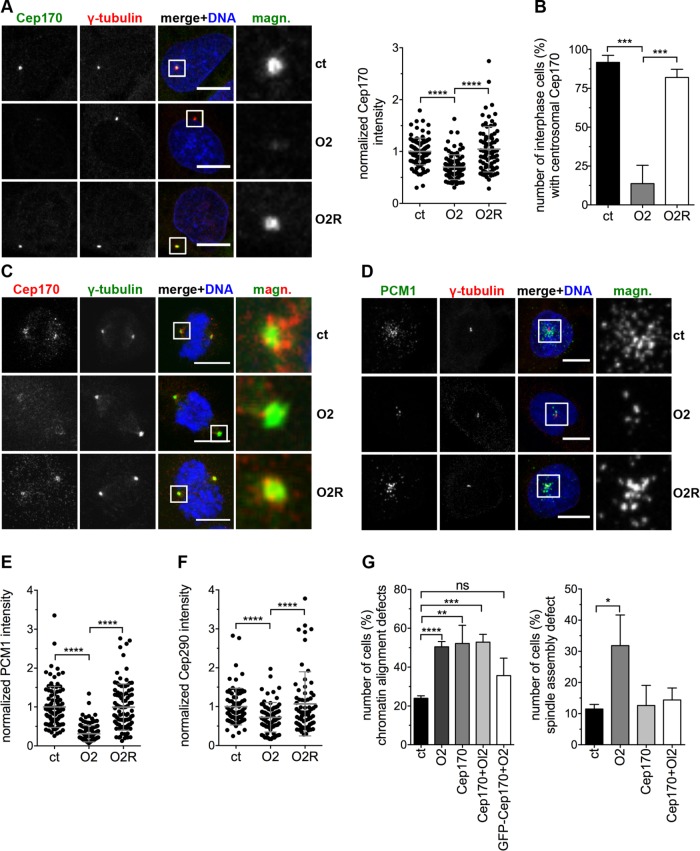
We anticipated that the observed MT defects after Ccdc61 depletion may be caused by mislocalization or deregulation of Cep170. To investigate this hypothesis, we performed Ccdc61 knockdown experiments in HeLa and U2OS cells and analyzed the localization of Cep170. Interestingly, Ccdc61 loss disturbed the anchorage or maintenance of Cep170 on centrosomes, leading to a significantly reduction of Cep170 at the centrosomes in interphase and mitotic cells (Figure 6, A–C, and Supplemental Figure S6, C–F). Cep170 can be efficiently reloaded by overexpression of a siRNA-resistant version of Ccdc61 (Figure 6, A–C); confirming the involvement of Ccdc61 in this process. Interestingly, similar effects were observed for the centriolar satellite protein PCM1. Depletion of Ccdc61 led to a significant reduction of its satellite localization, that could also been rescued after the overexpression of the siRNA-resistant Ccdc61 (Figure 6, D and E, and Supplemental Figure S7A). The localization of the satellite protein Cep290 depends on PCM1 (Kim et al., 2008). Thus, as expected, the satellite localization of Cep290 was also significantly reduced after loss of Ccdc61 (Figure 6F and Supplemental Figure S7, B–D). These results show, that localization of Cep170 and PCM1 depends on the centrosomal protein Ccdc61.
FIGURE 6:
Ccdc61 regulates the centrosomal anchorage of the MT binding protein Cep170. (A) U2OS cells were transfected with the indicated siRNAs or siRNA/plasmid combinations and the localization of Cep170 analyzed. Representative images show staining with Cep170 (green) and γ-tubulin (red); DNA was costained with Hoechst33258 (blue). Scattered dot plot shows the Cep170 intensity quantified in a 25-µm2 circle around the centrosome normalized to the control (ct). Data represent mean ± SD from three independent experiments, n > 77 cells. ****p < 0.0001 (unpaired Student’s t test). (B) Centrosomal localization of Cep170 in interphase U2OS cells after indicated treatments was determined by visual inspection of 120–150 cells per treatment and experiment. Data represent mean ± SD from three independent experiments, ***p < 0.001 (unpaired Student’s t test). (C) Mitotic U2OS cells were transfected with the indicated siRNAs or siRNA/plasmid combinations. γ-tubulin was used to visualize mitotic centrosomes (green), Cep170 visualized in red, and DNA in blue (Hoechst33258). Magnified panels (magn.) show enlarged views of the boxed regions. Bar, 10 µm. (D, E) U2OS cells were transfected with the indicated siRNAs or siRNA/plasmid combinations and the PCM1 localization analyzed. IF images in D show staining with PCM1 (green) and γ-tubulin (red); costaining of the DNA with Hoechst33258 (blue). Bar, 10 µm. Scattered dot plot in E shows the PCM1 intensity quantified in a 25-µm2 circle around the centrosome normalized to the ct. Data represent mean ± SD from three independent experiments, n > 92 cells. ****p < 0.0001 (unpaired Student’s t test). (F) U2OS cells were transfected with the indicated siRNAs or siRNA/plasmid combinations and the Cep290 levels around the centrosome (25-µm2 circle) quantified. Control (ct)-treated cells were normalized to 1.0. Data represent mean ± SD from three independent experiments, n > 72 cells. ****p < 0.0001 (unpaired Student’s t test). Representative images are in Supplemental Figure S7B. (G) U2OS cells stably expressing tagged α-tubulin and H2B were imaged every 5 min for 15–19 h 72 h after transfection of ct, O2, Cep170 siRNAs, a combination of O2 and Cep170 siRNAs (Cep170 + O2) or a combination of GFP-Cep170 together O2 (GFP-Ccdc61 + O2). Quantification shows the analyzed spindle assembly defects and chromatin alignment defect. Spindle assembly defects include various problems of the cell to assembly a properly formed spindle (including, e.g., multipolar spindle but also bipolar spindles struggling to align DNA as indicated in Figure 2, A and B, for O2 treatment). Chromatin alignment defects include disturbed formation of a properly arranged metaphase plate, e.g. lagging chromosomes. Data represent mean ± SD from three independent experiments, n > 135 cells. ns: not significant; *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001 (unpaired Student’s t test).
In a next step, we analyzed whether the spindle phenotypes observed after ablation of Ccdc61 depend on Cep170 (Figure 2C). Therefore, we performed siRNA-mediated down-regulation experiments of Cep170 alone or in combination with Ccdc61. Analysis of chromatin alignment in live cell experiments reveal that loss of either Cep170 or Cep170 together with Ccdc61 lead to a similar phenotype as ablation of Ccdc61 alone, resulting in a high number of cells showing chromatin alignment defects (Figure 6G and Supplemental Figure S7E). To further confirm the Cep170 dependency of Ccdc61, we analyzed whether overexpression of Cep170 could rescue chromatin alignment defects. Indeed, simultaneous down-regulation of Ccdc61 together with overexpression of GFP-Cep170 could partially rescue chromatin alignment defects (Figure 6G). Surprisingly, mitotic spindle assembly was not altered after siRNA-mediated down-regulation of Cep170 (Figure 6G). Altogether, our findings suggest that chromatin alignment seems to depend on both Ccdc61 and its downstream interaction partner Cep170, whereas proper mitotic spindle alignment might only require Ccdc61 function.
Ccdc61 promotes the complex formation of Cep170 and TBK1 that is required for MT stability
On the basis of the observed perturbation of the cellular distribution of Cep170 after depletion of Ccdc61, we were wondering how Ccdc61 maintains the centrosomal localization of Cep170. To address the possibility that Ccdc61 recruits Cep170 to its centrosomal localization, we performed fluorescence recovery after photobleaching (FRAP). We transiently expressed GFP-Cep170 in control or Ccdc61 siRNA-treated HeLa cells and analyzed the dynamic and mobility rate of GFP-Cep170. The Cep170 centrosomal recovery after photobleaching in unperturbed and Ccdc61 siRNA-treated cells was almost equal in all tested conditions (Supplemental Figure S7F), indicating that the centrosomal mobility and the recruitment of Cep170 are independent of Ccdc61. This observation further supports a role of Ccdc61 as an anchorage factor for the stability of Cep170.
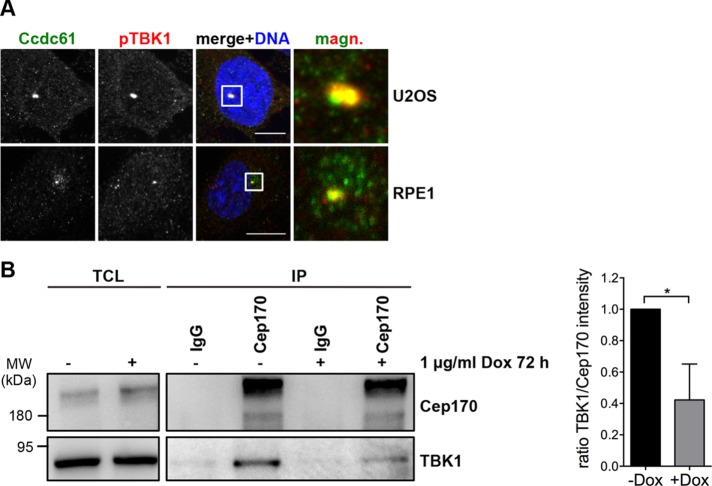
Previous findings revealed that Cep170 and TBK1 form a complex and that the interaction between both proteins is important for proper MT stability, mitotic spindle formation and mitosis (Pillai et al., 2015). The fact that TBK1 phosphorylates Cep170 and is essential for Cep170 centrosomal localization may point to a functional interplay among Ccdc61, Cep170, and TBK1. Interestingly, TBK1 also appeared in the Ccdc61 BioID approach, indicating its presence in close proximity to Ccdc61 (Supplemental Figure S5D). Although we were unable to detect a direct interaction between Ccdc61 and TBK1 in coimmunoprecipitation experiments, we observed a clear colocalization between the two proteins at the centrosome (Figure 7A). As Ccdc61 binds to Cep170 (Figure 5, A and C), we investigated whether complex formation between Cep170 and TBK1 could be impaired on Ccdc61 depletion. In fact, we found that the Cep170-TBK1 binding was markedly decreased when Ccdc61 was down-regulated (Figure 7B). As binding and phosphorylation of Cep170 by TBK1 is required for the centrosomal localization of Cep170, our data suggest that Ccdc61 plays an import role as a mediator of this interaction.
FIGURE 7:
Ccdc61 promotes the complex formation of Cep170 with TBK1. (A) IF analysis of U2OS and RPE1 cells in interphase. Images show endogenous localization of Ccdc61 (green) and phospho-TBK1 (pTBK1) in red visualized with specific antibodies. Chromatin was visualized by Hoechst33258 staining (blue). Bar, 10 µm. (B) Endogenous immunoprecipitation experiment using HeLa S/A Ccdc61 miRNA Oligo2 tet on cells. Cells were either untreated (–) or Ccdc61 miRNA O2 expression was induced via incubation with 1 µg/ml doxycycline for 72 h (+). Immunoprecipitation was performed using normal mouse IgG or mouse anti-Cep170 antibody coupled to ProteinG beads. Western blot shows TCL as input control and IP fractions. Mouse anti-Cep170 antibody was used to visualize endogenous Cep170 and anti-TBK1 antibody was used for detection of endogenous TBK1. Quantification shows the normalized ratio of TBK1/Cep170 intensity. Data represent mean ± SD from three independent experiments, *p < 0.05 (unpaired Student’s t test).
DISCUSSION
Here, we demonstrated that the so far uncharacterized centrosomal protein Ccdc61 affects mitosis, including MT organization, spindle assembly, chromatin alignment and spindle symmetry (Figures 2–4). Resulting defects on the mitotic spindle after ablation of Ccdc61 thereby lead to a significant delay in mitotic progression (Figure 2A).
A BioID approach for proteins identified in close proximity to Ccdc61, including PCM1, Cep290, Cep170, and TBK1, is known to be directly involved in MT anchoring and organization as well as in ciliogenesis and mitosis (Supplemental Figure S5) (Guarguaglini et al., 2005; Pillai et al., 2015; Hori and Toda, 2017). These proteins identified as novel complex components with Ccdc61 well explain the functional role of Ccdc61 in mitotic progression and MT organization.
Centriolar satellite proteins have an important mitotic function in spindle pole assembly, MT anchorage and chromatin alignment (Tollenaere et al., 2015). These include Cep72, Cep90, hMsd1/SSX2IP, and WDR8. Cep72 and Cep90 are required for maintaining spindle pole stability and chromosome alignment during metaphase through the recruitment of Kizuna and γ-tubulin (Oshimori et al., 2009; Kim and Rhee, 2011; Kim et al., 2012; Luddecke et al., 2016). hMsd1/SSX2IP together with its interaction partner WDR8, were shown to function in the correct anchorage of MTs to the centrosome. Also, the knockdown of WDR8 or hMsd1/SSX2IP displayed very similar mitotic defects, in which spindle MTs became shortened and misoriented (Barenz et al., 2013; Hori et al., 2014, 2015a,b). For these satellites structures, PCM1 is thought to function as a scaffold for other proteins to reside within the satellites. Consequently, depletion of PCM1 results in a disassembly of satellite structures (reviewed in [Hori and Toda, 2017]). Although Ccdc61 exhibits a satellite-like, dispersed staining pattern around centrosomes and also partially colocalizes with PCM1, we found that PCM1-depletion does not lead to disassembly of Ccdc61 staining around centrosomes (Figure 1 and Supplemental Figure S2C). Ccdc61 might therefore not be a bona fide centriolar satellite protein. However, it could still be that PCM1 influences Ccdc61 integrity or interacts with Ccdc61 in a cell cycle–dependent manner, since both protein change their localization throughout the cell cycle (Figure 1) (Hori et al., 2014). Interestingly, our findings (Figure 6, D and E, and Supplemental Figure S7A) implicate that the localization of PCM1 at centrosomes is dependent on Ccdc61. Thus, it is conceivable that Ccdc61 might regulate satellite function through PCM1 and might therefore also transiently localize to satellites.
Cep290 is a centriolar satellite component that apart from its function in ciliogenesis is also involved in mitotic progression and spindle positioning (Song et al., 2015). Interestingly, Ccdc61 depletion affects the correct loading of Cep290 on mitotic centrosomes (Figure 6F and Supplemental Figure S7, B–D). The proper localization of single centriolar satellite components is very often necessary for the correct localization of other satellite factors (Tollenaere et al., 2015). This interdependence seems to be true for Ccdc61 and Cep290.
Cep170 itself is partially associated with subdistal appendages (Guarguaglini et al., 2005) and our data indicated that Ccdc61 colocalizes to Cep170 in the vicinity of these structures (Figure 5D and Supplemental Figure S6A). Although proteins located at the subdistal appendage have been implicated in various processes, subdistal appendages seem to be important sites for MT anchoring (Bornens, 2002). Therefore, it is plausible that Cep170 and Ccdc61 work in concert to control MT anchoring to centriolar appendages, which in turn contributes to the correct organization of the MT network within the cell. Cep170 is additionally necessary to target Kif2b to the mitotic spindle and can thereby impart its MT binding activity to Kif2b and Kifc3 locating these kinesins to the mitotic spindle (Welburn and Cheeseman, 2012). Additionally, it was shown that Cep170 forms a complex with TBK1, which is also required for Cep170 interplay with Kif2b and that loss of TBK1 alters MT stability and inhibits mitosis (Pillai et al., 2015). Since Ccdc61 alters complex formation of Cep170 and TBK1, it could well be that Ccdc61 is another player in this complex regulation. Thus, Ccdc61 might stimulate mitotic processes and MT organization via Cep170/TBK1 and might also indirectly impair the correct MT targeting of Kif2b.
In summary, we have discovered Ccdc61 as novel protein regulating centriolar satellite components with a critical function in mitotic MT organization and mitotic progression through the correct centrosomal maintenance of Cep170.
MATERIAL AND METHODS
Cell culture, generation of stable Ccdc61 expressing cell line, generation of a stable tet-on Ccdc61 miRNA cell line, transfection of plasmids, or siRNAs and rescue experiments
HeLa (ATCC CCL-2), HeLa S/A (O. Gruss, University of Bonn, Germany), U2OS (ATCC HTB-96), HeLa α-tubulin–mEGFP mCherry-H2B (D. Gerlich, IMBA, Austria), U2OS H2B-GFP α-tubulin–mCherry (S. Geley, Innsbruck Medical University, Austria), HEK293T (ACC 635; DSMZ, Braunschweig, Germany), HEK293T Flp-In-T-Rex (M. Lemberg, ZMBH, Heidelberg, Germany), HeLa-TDS-mouse Ccdc61 (mCcdc61)-LAP (A. Hyman, MPI, Dresden, Germany) cells were grown in high-glucose DMEM (Life Technologies) and hTERT-RPE1 cells (RPE1; ATCC, CRL-4000) in DMEM/F12 (50:50, Life Technologies), supplemented with 10% fetal bovine serum (FBS) (Sigma-Aldrich) and 1 mM l-glutamine (Sigma-Aldrich). HeLa S/A cells stably expressing Ccdc61 miRNA Oligo2 under the control of a tetracycline inducible promoter were generated. The expression of the miRNA was induced by the addition of 1 µg/ml doxycycline. HEK293T Flp-In T-Rex FLAG-Ccdc61 (rescue)-BirA* was generated according to the manufacturer’s instructions (Life Technologies) and expression induced by addition of 1 µg/ml doxycycline. Cell lines were periodically tested for contaminations and cell line authenticated by Multiplexion, Heidelberg, Germany.
Transfections were performed with Lipofectamine 2000 or Lipofectamine RNAiMax (Life Technologies), using 1–3 µg of plasmid and/or 20–40 nM siRNA to transfect six-well plates according to the manufacturer’s recommendations. To perform immunoprecipitation experiments, HEK293T cells were transfected with polyethylenimine (Polysciences) at a final concentration of 5 µg/ml using 25 µg plasmid DNA per 15 cm2 dish (2 × 107 cells). Cells were synchronized at the G1/S boundary using a thymidine block. Cells were thereby arrested by the addition of 2–3 mM thymidine (Sigma-Aldrich) for 19–24 h and afterward released by five washes with phosphate-buffered saline (PBS). For rescue experiments, Ccdc61 siRNA Oligo2 resistant was transiently transfected into U2OS or HeLa cells either at the same time as the siRNA or 48 h after siRNA transfection and expressed for 24–72 h. After 72 h, cells were fixed and processed for IF analysis. To depolymerize MTs, cells were either incubated for 45 min on ice and reincubated at 37°C to or incubated for 2 h with 20 µM nocodazole (AppliChem) at 37°C, and afterward washed three times with PBS and reincubated at 37°C to allow MT regrowth.
Antibodies
Recombinant full-length human Ccdc61 protein was used for monoclonal antibody generation in mice by the DKFZ monoclonal antibody core facility. The resulting monoclonal mouse anti-Ccdc61 antibody was affinity purified using Ccdc61 protein immobilized on CNBr-activated Sepharose 4B (GE Healthcare) according to manufacturer’s recommendations, and used at a final concentration of 50–100 ng/µl. Mouse anti-FLAG M2 (1:5000; F3165), mouse anti-α-tubulin (1:1000; T5168) and mouse anti-γ-tubulin (1:500; T6557) were obtained from Sigma, goat anti-γ-tubulin C-20 (1:50, sc-7396) from Santa Cruz, rabbit anti-GFP (1:500; NB600-303) from Novus, rabbit anti-pericentrin (1:3000; ab4448) and rabbit anti-Cep170 (1:500; ab72505) from Abcam, mouse anti-Cep170 72-413-1 (1:1000 or 1 µg per 1 mg total cell lysate; 41-3200) from Thermo Scientific, rabbit anti-Cep290 (1:500; A301-659A-2) from Bethyl and rabbit anti-Mad2 (1:100; PRB-452C) from Covance. Mouse Penta-His (1:300; 34660) was obtained from Qiagen and mouse anti-actin Ab1 (1:5000; JLA20) from Millipore. Rabbit anti-TBK1/NAK D1B4 (1:1000, 3504) and rabbit anti-phospho-TBK1/NAK (1:50, 5483) from Cell Signaling, rabbit ODF-2 (1:500, 12058-1-AP) from Proteintech, and human anti-CREST (3600; 1:5000). Rabbit anti-PCM1 (1:1000) was provided by A. Merdes (Université de Toulouse, Toulouse, France), goat anti-C-Nap1 (1:500) provided by E. Schiebel (ZMBH, Heidelberg, Germany) and rat anti-ninein (1:500) provided by G. Chen (University of Alberta, Edmonton, Alberta, Canada). Normal mouse IgG 1 (SC2025) was obtained from Santa Cruz. If possible, antibodies were validated by RNAi experiments with Western blot or IF.
Secondary antibodies for IF were goat anti-mouse IgG and goat anti-rabbit IgG coupled to Alexa Fluor 488 and Alexa Fluor 594 (1:500–1:1000; Molecular Probes) or donkey anti-mouse immunoglobulin G (IgG), donkey anti-rabbit IgG, and donkey anti-goat IgG coupled to Alexa Fluor 488, Alexa Fluor 568 or Alexa Fluor 647 (1:500–1:1000, Molecular Probes). Secondary antibodies for Western blotting were peroxidase-conjugated donkey anti-rabbit (1:5000–1:10,000; Jackson Laboratories), goat anti-mouse (1:5000–1:10,000; Novus), and Streptavidin-HRP (1:2000, Thermo Scientific 815-968-0747).
siRNAs
siRNAs used in this study were directed against the following sequences: firefly luciferase (GL2, control), 5′-AACGUACGCGGAAUACUUCGA-3′; Ccdc61 Oligo1, 5′-CCCUGGAGUGGGUCCAAUA-3′; Ccdc61 Oligo2, 5′-AGAUUUGACUCACAAGACA-3′; Cep170 5’-GCAUGAGAAGUUUACCAUU-3′; PCM1 5′-GTATCACATCTGAACTAAA-3′. All siRNAs were purchased from Eurofins Genomics, Ebersberg, Germany, at high-performance liquid chromatography purity.
Plasmid constructions and molecular cloning
Full-length human Ccdc61 cDNA (NCBI Gene ID: 729440) was synthesized by Geneart/Life Technologies and cloned via PCR amplification into the BamHI/HindIII sites of pCMV-3Tag1A (Agilent Technologies), into the HindIII/SalI sites of pEGFP-C3 (Clonetech), into the BamHI/HindIII sites of pET22b (Merck Millipore), into the HindIII/BamHI sites of pcDNA5-FLAG-BirA*-R118G (provided by L. Pelletier, Department of Molecular Genetics University of Toronto), and into the KpnI/ApaI sites of pcDNA5 FRT TO (Life Technologies) using pCMV-3Tag-1A-Ccdc61 as a template.
siRNA-resistant human Ccdc61 was generated by PCR-based site-directed mutagenesis (QuikChange Lightning Multi-Site directed Mutagenesis Kit, Agilent), mutating residues pairing to the seed region of Oligo2 from 5′-GATTTGACTCACAAGACA-3′ to 5′-GACCTAACGCATAAAACA-3′ introducing silent mutations within the protein encoding sequence. pCMV-3Tag-1A-Ccdc61 was used as a template. DNA sequences were analyzed by sequencing.
Recombinant protein expression
Human Ccdc61-6xHis was expressed in Escherichia coli BL21-Rosetta and purified under denaturing conditions by single-step affinity chromatography using Ni-NTA Sepharose (Qiagen) according to the instructions of the manufacturer. Ccdc61 was finally dialyzed against PBS, pH 7.4 containing 8 M urea.
Immunoprecipitation
HEK293T cell lysates were prepared with NP40 lysis buffer (40 mM Tris, pH 7.5, 150 mM NaCl, 0.5% NP40, 5 mM EDTA, 10 mM β-glycerophosphate, 5 mM NaF, 1 mM dithiothreitol, 0.1 mM Na3VO4, and protease inhibitors). Immunoprecipitations were performed as shown previously (Zhu et al., 2013).
BioID and mass spectrometry analysis
HEK293T Flp-In T-Rex FLAG-Ccdc61 (rescue)-BirA* cells were induced with doxycycline (1 μg/ml, 24 h) and treated with or without biotin (50 μM, 24 h; Sigma-Aldrich). Cells were harvested and afterward washed twice in PBS. Biotin-streptavidin affinity purification from total cell extracts was performed as described previously (Firat-Karalar and Stearns, 2015). Mass spectrometry analysis of the samples was performed as described previously (Kratz et al., 2015).
IF of fixed specimens
For indirect IF HeLa, U2OS, HEK293T, and RPE1 cells were treated as described previously (Cizmecioglu et al., 2010; Kschonsak and Hoffmann, 2018). Coverslips were mounted onto glass slides with ProLong Gold (Molecular Probes) or Mowiol (Calbiochem). For cell imaging, the Zeiss motorized inverted Observer.Z1 microscope was used, containing mercury arc burner HXP 120 C and LED module Colibri. Filter combinations: GFP (38 HE) DsRed (43 HE), and 4′,6-diamidino-2-phenylindole (49) with the detector gray-scale charge-coupled device (CCD) camera AxioCam MRm system and a 63 ×/1.4 Oil Pln Apo DICII objective. Zeiss Apotome confocal imaging device was used during z-stack imaging. For quantitative or colocalization purpose, cells were imaged with the Zeiss LSM 710 confocal microscope, with a 63×/1.4 oil DIC III objective, and containing laser diodes 405-5, 488, 555, and 633. Image processing was performed using ZEN and Fiji (Schindelin et al., 2012), including maximum z-stack projection and background subtraction.
Centrosomal or satellite localization of PCM1, Cep290, Cep170, and Ccdc61 were quantified by randomly chosen cells by drawing a 3- to 25-µm2 circle around the centrosome and measuring the mean gray intensity of the corresponding signal. MT aster size was measured my drawing a circle with the centrosome centered accordingly to the length of the MTs. Colocalization of Ccdc61 with PCM1 or pericentrin was quantified using a Fiji macro (Schindelin et al., 2012). Thereby, a mask of the specific PCM/pericentrin signal of a cell was generated and transferred to the Ccdc61 signal to measure the intensity of Ccdc61 in this specific area.
Live cell imaging
U2OS cells stably expressing H2B-GFP α-tubulin–mCherry or HeLa cells stably expressing α-tubulin–mEGFP mCherry-H2B were used for time-lapse experiments. Cells were treated with the indicated siRNA, synchronized via thymidine block, seeded into a µ-slide 8-well ibiTreat chambers (ibidi GmbH) and imaged for 16–19 h at a 5 min time interval on a Zeiss motorized inverted Observer.Z1 microscope, containing mercury arc burner HXP 120 C and LED module Colibri. UPLS-Apochromat 20 ×/0.75 NA air objective lens was used for imaging and the Zeiss ZEN software for image acquisition. Filter combinations: GFP (38 HE) and DsRed (43 HE) with the detector gray scale CCD camera AxioCam MRm system. Cells were kept in DMEM/FBS at 37°C and 5% CO2 during imaging. Images were analyzed with Fiji (Schindelin et al., 2012). Cells were randomly chosen and the timing of mitosis were analyzed by measuring the time between nuclear envelope breakdown and telophase (mitotic duration) or mitotic spindle formation (metaphase) and anaphase. Chromatin alignment defects were analyzed by using the H2B channel only. Spindle assembly defects include various problems of the cell to assembly a properly formed spindle (including, e.g., multipolar spindle but also bipolar spindles struggling to align DNA). Chromatin alignment defects include disturbed formation of a properly arranged metaphase plate, for example, lagging chromosomes.
Fluorescence recovery after photobleaching (FRAP) analysis
FRAP measurements were performed on a Zeiss LSM 780 confocal microscope using a 40×/1.3 numerical aperture (NA) oil Plan-Apochromat objective, the 488-nm line of an Argon laser and open pinhole settings. Cells were grown on eight-well µ-slides (ibidi) and subjected to FRAP analyses for no longer than 1 h on the microscope at 37°C. For quantification, recovery curves were derived from six cells investigated for each condition. The mobile fraction was calculated by comparing the photobleach corrected prebleach and postbleach recovery fluorescence intensity values in the photobleached region of interest. Image processing and analysis was performed using Fiji (Schindelin et al., 2012).
Live-cell imaging of MT plus ends
HeLa EGFP-EB3-expressing cells (Sironi et al., 2011) were seeded into eight-well µ ibidi chambers (ibidi GmbH, Germany). The cells were imaged at a Leica TCS SP5 confocal microscope supported with a temperature and CO2-controlled incubation chamber using HCX Plan APO 40×/1.30 Oil CS objective and the Leica Application Suite at a temporal resolution of 0.13 s. siR-DNA dye was added to the cells to visualize the DNA.
Tracking of MTs in mitosis and quantitative analysis of MT dynamics
The goal was to quantify the directionality and asymmetry of many mitosis events; each with many closely spaced tracks, monitored over arbitrarily large time intervals. It was especially important to decouple parameters such as position, size, and absolute orientation of the spindle for meaningful and easy comparison of different mitosis events.
At the core of our technique was the tracking program TrackMate (http://imagej.net/TrackMate), which is part of the image analysis program Fiji (Schindelin et al., 2012). We first optimized four major parameters of this program (spot diameter, intensity threshold, maximum displacement of a spot from one frame to the next, and the minimum acceptable duration of a track) for a typical time-lapse image series. These parameters were then used consistently for automated tracking of all time-lapse images (script-based use of TrackMate) and obtaining tracks.
A detected track included links that join a given MT tip from one frame to the next. Knowing the positions of the MT tips, the directions of the links/tracks could then be easily calculated. The histogram of these directions was used to quantify the directionality and asymmetry indices, as shown in Supplemental Figure S4C. The horizontal axis represents angles (in degrees), and the vertical axis represents the number of detected links (two consecutive spots along a given track) per unit of angle. The two (Orientation and Biorientation) indices quantify the directionality and the asymmetry of microtubule growths, irrespective of microtubule spindle size, position, or orientation. As such, they make it possible to compare different mitosis events automatically without any registration, scaling, or region of interest selection. Directionality was defined as the area under the histogram around the dominant orientation down to the baseline (A1) divided by the corresponding area under the baseline (A0). Asymmetry was defined as the normalized difference between excess numbers of links growing along the dominant orientation (A1) and the rest of the links (A2).
Tracking particles in a dense medium required small displacement of a given spot (from one frame to the next) compared with its distance to adjacent spots (in a single frame). To this end, we used high-frame-rate imaging (0.13 s/frame) and also reduced laser power to avoid acquisition bleaching. The resulting reduced signal-to-noise ratio was no problem for the robust tracking algorithm, as verified by visual inspection in several cases. More details of the technique, its open-source implementation, and its robustness to many experimental and computational challenges are addressed in a different contribution. More details of the technique, its open-source implementation, and its robustness to many experimental and computational challenges are addressed in a preprint (Jafarpour and Lorenz, 2017).
Statistical analysis
Statistical analysis was performed with GraphPad Prism 6.0. The results shown in the figures represent the mean ± SD, if not indicated otherwise. Unpaired Student’s t test (two-tailed distribution), one-way analysis of variance (ANOVA), or two-way ANOVA was performed to calculate the p value that is indicated with asterisks or as ns (not significant). Statistical details of each experiment can be found in the figure legends. No statistical method was used to predetermine sample size.
Supplementary Material
Acknowledgments
We thank A. Merdes for providing PCM1 antibody and L. Pelletier for providing the pcDNA5-FLAG-BirA*-R118G-plasmid. We are grateful to M. Lemberg and O. Gruss for providing the T-Rex cell lines and HeLa S/A cell line, D. Gerlich for providing the HeLa α-tubulin–mEGFP mCherry-H2B, cell line, and S. Geley for providing the U2OS H2B-GFP α-tubulin–mCherry. G. Guarguaglini is thanked for providing Cep170 fragment versions. We acknowledge K. Johansson for providing SiR-DNA, G. Chen for the anti-ninein antibody, and E. Schiebel for the anti-C-Nap1 antibody. This work was supported by a grant from the Deutsche Krebshilfe (110243) and from the Deutsche José Carreras Leukämie-Stiftung (DJCLC 09/30f), both to I.H.
Abbreviations used:
- Ccdc61
coiled-coil containing protein 61
- Cep170
centrosomal protein of 170 kDa
- ct
control
- FRAP
fluorescence recovery after photobleaching
- H2B
histone 2B
- IF
immunofluorescence
- MT
microtubule
- O1
siRNA #1 directed against Ccdc61 (Oligo1)
- O2
siRNA #2 directed against Ccdc61 (Oligo2)
- O2R
siRNA #2 directed against Ccdc61 transfected together with siRNA-resistant FLAG-Ccdc61 (rescue construct)
- PCM
pericentriolar material
- PCM1
pericentriolar material 1 protein
- SAC
spindle assembly checkpoint
- TBK1
TANK binding kinase 1.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E18-02-0115) on October 24, 2018.
REFERENCES
- Applegate KT, Besson S, Matov A, Bagonis MH, Jaqaman K, Danuser G. (2011). plusTipTracker: Quantitative image analysis software for the measurement of microtubule dynamics. J Struct Biol , 168–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balczon R, Bao L, Zimmer WE. (1994). PCM-1, A 228-kD centrosome autoantigen with a distinct cell cycle distribution. J Cell Biol , 783–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balczon R, Simerly C, Takahashi D, Schatten G. (2002). Arrest of cell cycle progression during first interphase in murine zygotes microinjected with anti-PCM-1 antibodies. Cell Motil Cytoskeleton , 183–192. [DOI] [PubMed] [Google Scholar]
- Barenz F, Inoue D, Yokoyama H, Tegha-Dunghu J, Freiss S, Draeger S, Mayilo D, Cado I, Merker S, Klinger M, et al (2013). The centriolar satellite protein SSX2IP promotes centrosome maturation. J Cell Biol , 81–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barenz F, Mayilo D, Gruss OJ. (2011). Centriolar satellites: busy orbits around the centrosome. Eur J Cell Biol , 983–989. [DOI] [PubMed] [Google Scholar]
- Bauer M, Cubizolles F, Schmidt A, Nigg EA. (2016). Quantitative analysis of human centrosome architecture by targeted proteomics and fluorescence imaging. EMBO J , 2152–2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornens M. (2002). Centrosome composition and microtubule anchoring mechanisms. Curr Opin Cell Biol , 25–34. [DOI] [PubMed] [Google Scholar]
- Bouckson-Castaing V, Moudjou M, Ferguson DJ, Mucklow S, Belkaid Y, Milon G, Crocker PR. (1996). Molecular characterisation of ninein, a new coiled-coil protein of the centrosome. J Cell Sci (Pt 1), 179–190. [DOI] [PubMed] [Google Scholar]
- Chinen T, Liu P, Shioda S, Pagel J, Cerikan B, Lin TC, Gruss O, Hayashi Y, Takeno H, Shima T, et al (2015). The gamma-tubulin-specific inhibitor gatastatin reveals temporal requirements of microtubule nucleation during the cell cycle. Nat Commun , 8722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chretien D, Buendia B, Fuller SD, Karsenti E. (1997). Reconstruction of the centrosome cycle from cryoelectron micrographs. J Struct Biol , 117–133. [DOI] [PubMed] [Google Scholar]
- Cizmecioglu O, Arnold M, Bahtz R, Settele F, Ehret L, Haselmann-Weiss U, Antony C, Hoffmann I. (2010). Cep152 acts as a scaffold for recruitment of Plk4 and CPAP to the centrosome. J Cell Biol , 731–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conduit PT, Wainman A, Raff JW. (2015). Centrosome function and assembly in animal cells. Nat Rev Mol Cell Biol , 611–624. [DOI] [PubMed] [Google Scholar]
- Dammermann A, Merdes A. (2002). Assembly of centrosomal proteins and microtubule organization depends on PCM-1. J Cell Biol , 255–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumoux M, Menny A, Delacour D, Hayward RD. (2015). A Chlamydia effector recruits CEP170 to reprogram host microtubule organization. J Cell Sci , 3420–3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firat-Karalar EN, Stearns T. (2015). Probing mammalian centrosome structure using BioID proximity-dependent biotinylation. Methods Cell Biol , 153–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarguaglini G, Duncan PI, Stierhof YD, Holmstrom T, Duensing S, Nigg EA. (2005). The forkhead-associated domain protein Cep170 interacts with Polo-like kinase 1 and serves as a marker for mature centrioles. Mol Biol Cell , 1095–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta GD, Coyaud E, Goncalves J, Mojarad BA, Liu Y, Wu Q, Gheiratmand L, Comartin D, Tkach JM, Cheung SW, et al (2015). A dynamic protein interaction landscape of the human centrosome-cilium interface. Cell , 1484–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori A, Ikebe C, Tada M, Toda T. (2014). Msd1/SSX2IP-dependent microtubule anchorage ensures spindle orientation and primary cilia formation. EMBO Rep , 175–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori A, Morand A, Ikebe C, Frith D, Snijders AP, Toda T. (2015a). The conserved Wdr8-hMsd1/SSX2IP complex localises to the centrosome and ensures proper spindle length and orientation. Biochem Biophys Res Commun. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori A, Peddie CJ, Collinson LM, Toda T. (2015b). Centriolar satellite- and hMsd1/SSX2IP-dependent microtubule anchoring is critical for centriole assembly. Mol Biol Cell , 2005–2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori A, Toda T. (2017). Regulation of centriolar satellite integrity and its physiology. Cell Mol Life Sci , 213–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H, Kubo A, Tsukita S, Tsukita S. (2005). Odf2-deficient mother centrioles lack distal/subdistal appendages and the ability to generate primary cilia. Nat Cell Biol , 517–524. [DOI] [PubMed] [Google Scholar]
- Jafarpour A, Lorenz H. (2017). Cellulyzer—automated analysis and interactive visualization/simulation of select cellular processes. arXiv:1703.02611v1 (physics.bio-ph). [Google Scholar]
- Jakobsen L, Vanselow K, Skogs M, Toyoda Y, Lundberg E, Poser I, Falkenby LG, Bennetzen M, Westendorf J, Nigg EA, et al (2011). Novel asymmetrically localizing components of human centrosomes identified by complementary proteomics methods. EMBO J , 1520–1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Krishnaswami SR, Gleeson JG. (2008). CEP290 interacts with the centriolar satellite component PCM-1 and is required for Rab8 localization to the primary cilium. Hum Mol Genet , 3796–3805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K, Lee K, Rhee K. (2012). CEP90 is required for the assembly and centrosomal accumulation of centriolar satellites, which is essential for primary cilia formation. PLoS One , e48196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K, Rhee K. (2011). The pericentriolar satellite protein CEP90 is crucial for integrity of the mitotic spindle pole. J Cell Sci , 338–347. [DOI] [PubMed] [Google Scholar]
- Klinger M, Wang W, Kuhns S, Barenz F, Drager-Meurer S, Pereira G, Gruss OJ. (2014). The novel centriolar satellite protein SSX2IP targets Cep290 to the ciliary transition zone. Mol Biol Cell , 495–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kratz AS, Barenz F, Richter KT, Hoffmann I. (2015). Plk4-dependent phosphorylation of STIL is required for centriole duplication. Biol Open , 370–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kschonsak YT, Hoffmann I. (2018). Activated ezrin controls MISP levels to ensure correct NuMA polarization and spindle orientation. J Cell Sci , jcs214544. [DOI] [PubMed] [Google Scholar]
- Kubo A, Sasaki H, Yuba-Kubo A, Tsukita S, Shiina N. (1999). Centriolar satellites: molecular characterization, ATP-dependent movement toward centrioles and possible involvement in ciliogenesis. J Cell Biol , 969–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo A, Tsukita S. (2003). Non-membranous granular organelle consisting of PCM-1: subcellular distribution and cell-cycle-dependent assembly/disassembly. J Cell Sci , 919–928. [DOI] [PubMed] [Google Scholar]
- Luddecke S, Ertych N, Stenzinger A, Weichert W, Beissbarth T, Dyczkowski J, Gaedcke J, Valerius O, Braus GH, Kschischo M, Bastians H. (2016). The putative oncogene CEP72 inhibits the mitotic function of BRCA1 and induces chromosomal instability. Oncogene , 2398–2406. [DOI] [PubMed] [Google Scholar]
- Mazo G, Soplop N, Wang WJ, Uryu K, Tsou MF. (2016). Spatial control of primary ciliogenesis by subdistal appendages alters sensation-associated properties of cilia. Dev Cell , 424–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meraldi P, Nigg EA. (2002). The centrosome cycle. FEBS Lett , 9–13. [DOI] [PubMed] [Google Scholar]
- Nakagawa Y, Yamane Y, Okanoue T, Tsukita S, Tsukita S. (2001). Outer dense fiber 2 is a widespread centrosome scaffold component preferentially associated with mother centrioles: its identification from isolated centrosomes. Mol Biol Cell , 1687–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshimori N, Li X, Ohsugi M, Yamamoto T. (2009). Cep72 regulates the localization of key centrosomal proteins and proper bipolar spindle formation. EMBO J , 2066–2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piel M, Meyer P, Khodjakov A, Rieder CL, Bornens M. (2000). The respective contributions of the mother and daughter centrioles to centrosome activity and behavior in vertebrate cells. J Cell Biol , 317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillai S, Nguyen J, Johnson J, Haura E, Coppola D, Chellappan S. (2015). Tank binding kinase 1 is a centrosome-associated kinase necessary for microtubule dynamics and mitosis. Nat Commun , 10072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prosser SL, Pelletier L. (2017). Mitotic spindle assembly in animal cells: a fine balancing act. Nat Rev Mol Cell Biol , 187–201. [DOI] [PubMed] [Google Scholar]
- Roux KJ, Kim DI, Raida M, Burke B. (2012). A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J Cell Biol , 801–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al (2012). Fiji: an open-source platform for biological-image analysis. Nat Methods , 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sironi L, Solon J, Conrad C, Mayer TU, Brunner D, Ellenberg J. (2011). Automatic quantification of microtubule dynamics enables RNAi-screening of new mitotic spindle regulators. Cytoskeleton (Hoboken) , 266–278. [DOI] [PubMed] [Google Scholar]
- Song H, Park JE, Jang CY. (2015). DDA3 targets Cep290 into the centrosome to regulate spindle positioning. Biochem Biophys Res Commun , 88–94. [DOI] [PubMed] [Google Scholar]
- Stowe TR, Wilkinson CJ, Iqbal A, Stearns T. (2012). The centriolar satellite proteins Cep72 and Cep290 interact and are required for recruitment of BBS proteins to the cilium. Mol Biol Cell , 3322–3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollenaere MA, Mailand N, Bekker-Jensen S. (2015). Centriolar satellites: key mediators of centrosome functions. Cell Mol Life Sci , 11–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welburn JP, Cheeseman IM. (2012). The microtubule-binding protein Cep170 promotes the targeting of the kinesin-13 depolymerase Kif2b to the mitotic spindle. Mol Biol Cell , 4786–4795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh C, Coyaud E, Bashkurov M, van der Lelij P, Cheung SW, Peters JM, Raught B, Pelletier L. (2015). The deubiquitinase USP37 regulates chromosome cohesion and mitotic progression. Curr Biol , 2290–2299. [DOI] [PubMed] [Google Scholar]
- Zhu M, Settele F, Kotak S, Sanchez-Pulido L, Ehret L, Ponting CP, Gonczy P, Hoffmann I. (2013). MISP is a novel Plk1 substrate required for proper spindle orientation and mitotic progression. J Cell Biol , 773–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.