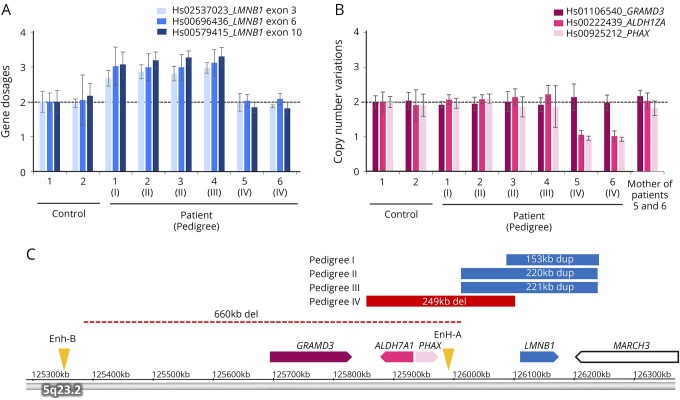
Figure 1. Analysis of CNVs of LMNB1 and its upstream region.
(A) Gene dosages for exons 3, 6, and 10 of LMNB1 were determined by TaqMan-based real-time PCR assay. The copy numbers of 3 exons of LMNB1 in patients 1–4 were increased by approximately 1.5-fold compared with control subjects, suggesting the presence of duplication of LMNB1 in these patients. (B) The copy number variations of regions upstream of LMNB1 including GRAMD3, ALDH7A1, and PHAX were determined by TaqMan-based real-time PCR assay. The copy numbers of ALDH7A1 and PHAX were decreased approximately by half in patients 5 and 6, suggesting the presence of the upstream deletion of LMNB1. (C) The genomic regions of duplication (blue) and deletion (red) were analyzed using an Affymetrix CytoScan HD array and are shown on the basis of information obtained from the UCSC genome browser (assembly GRCh37/hg19). The regions of duplication were 153 kb in pedigree I, 220 kb in pedigree II, and 221 kb in pedigree III. The deletion upstream of LMNB1 in a previous report is shown by a dotted line.8 The positions of original enhancer A (Enh-A) and alternative enhancer B (Enh-B) for LMNB1 are indicated by arrowheads. ALDH7A1 = aldehyde dehydrogenase 7 family member A1; CNV = copy number variation; GRAMD3 = GRAM domain containing 3; LMNB1 = lamin B1; PHAX = phosphorylated adaptor for RNA export.