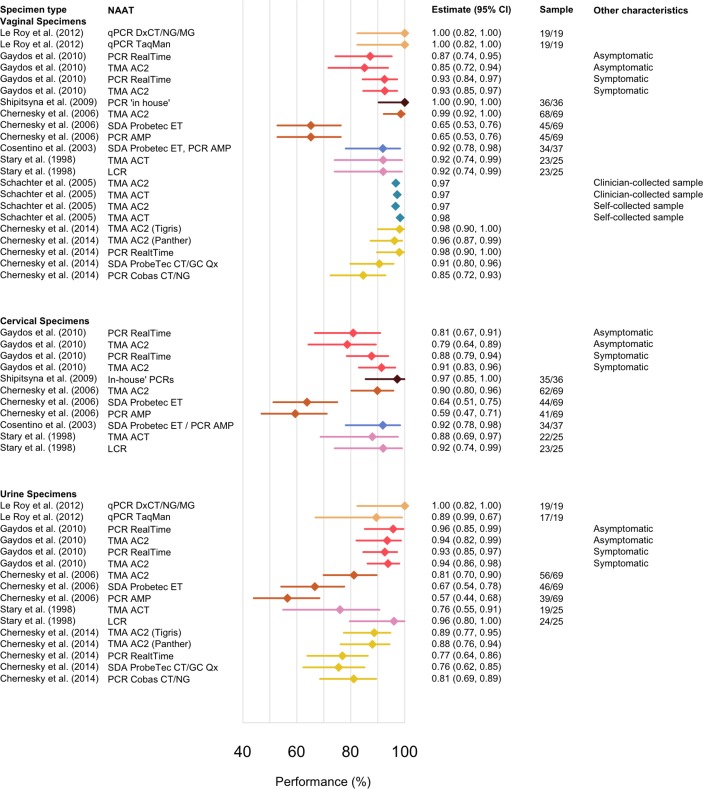
Figure 2.
Performance estimates for chlamydia by showing the sensitivity of the nucleic acid amplification test (NAAT) on a given specimen site relative to PIS. The studies are referenced by the first author, and publication year, and colours are by study for clarity. Each study is represented by a colour and where results are stratified by test, sampling or symptom status, this has been included in the figure. Tests used: LCR (ligase chain reaction); PCR AMP (PCR Ampicor); PCR Cobas CT/NG (PCR Cobas CT/NG); PCR ‘In house’; PCR RealTime (PCR RealTime); qPCR TaqMan (quantitative PCR TaqMan); qPCR DxCT/NG/MG (quantitative PCR DxCT/NG/MG); PCR LightMix, (PCR Lightmix); NASBA (nucleic acid sequence-based amplification); SDA ProbeTec ET (strand displacement amplification Probetec ET); SDA ProbeTec CT/GC Qx (strand displacement amplification ProbeTec CT/GC Qx); TMA AC2 (transcription-mediated amplification Aptima Combo-2); TMA ACT (transcription-mediated amplification Aptima CT); TMA AGC (transcription-mediated amplification Aptima GC).