Abstract
Urothelial carcinoma of the bladder is one of the most common malignancies in the industrialized world, mainly caused by smoking and occupational exposure to chemicals. The favorable prognosis of early stage bladder cancer underscores the importance of early detection for treatment of this disease. The high recurrence rate of this malignancy also highlights the need for close post-diagnosis monitoring of bladder cancer patients. As for other malignancies, aberrant DNA methylation has been shown to play a crucial role in the initiation and progression of bladder cancer, and thus holds great promise as a diagnostic and prognostic biological marker. Here, we describe a protocol for a versatile DNA methylation enrichment method, the Methylated CpG Island Recovery Assay (MIRA), which enables analysis of the DNA methylation status in individual genes or across the entire genome. MIRA is based on the ability of the methyl-binding domain (MBD) proteins, the MBD2B/MBD3L1 complex, to specifically bind methylated CpG dinucleotides. This easy-to-perform method can be used to analyze the methylome of bladder cancer or urothelial cells shed in the urine to elucidate the evolution of bladder carcinogenesis and/or identify epigenetic signatures of chemicals known to cause this malignancy.
Keywords: aromatic amines, biomarkers, epigenetics, methyl-binding domain (MBD) proteins, tobacco smoke, urine
1. INTRODUCTION
Bladder cancer is the ninth most common cancer in the world, with an estimated 430,000 new cases and 165,000 deaths in 2012 [1–3]. In the US alone, 76,960 new cases are expected to occur in 2016, and 16,390 bladder cancer patients are expected to die from the disease in the same year [4]. The vast majority (>90%) of bladder cancer cases are urothelial carcinomas. Approximately 75% of patients are diagnosed with non-muscle-invasive bladder cancer, of which roughly 50% are of low-grade [1, 3]. The prognosis of these non-invasive tumors is usually favorable; however, up to 80% of cases will recur after complete transurethral resection, and up to 45% of cases will progress to invasive cancer within 5 years [5, 6]. Cystoscopy, followed by biopsy of suspicious lesions, remains the gold standard for detection of both new and recurrent bladder cancer. However, this approach is highly invasive and costly and its sensitivity can be as low as 60% for detection of carcinoma in situ [6, 7]. Non-invasive tests for bladder cancer diagnosis are available and include voided urine cytology, cytogenetic analysis by fluorescence in situ hybridization, and detection of genetic mutations in urine. Yet, these tests have limited sensitivities (54–86%), and specificities (61–90%), and often yield false positive results [3, 6, 8]. Given the high recurrence rate of bladder cancer and the costs and discomfort associated with post-diagnosis follow-ups, the quest for novel non-invasive tests to improve early detection and assist with surveillance is currently a top research priority [8, 9].
Unlike other types of human cancer with unknown or less well-defined etiologic agent(s), bladder cancer is primarily linked to exposure to aromatic amines, a family of chemicals present in tobacco smoke and various industrial products and workplace settings [10–13]. Epidemiologic studies have shown that smokers have 2- to 6-fold higher risk of bladder cancer relative to non-smokers [1, 14]. Also, those who smoke black (air-cured) tobacco products are at increased risk of bladder cancer compared to those who smoke blond tobacco (flue-cured) products. The latter is consistent with the higher content of aromatic amines in smoke from black tobacco products relative to blond tobacco products [11]. Furthermore, higher incidence of bladder cancer has been observed in industrial workers exposed to aromatic amines generated during the manufacture or processing of a variety of products, including rubber, cable, textile, dye, paints, solvents, leather dust, inks, etc [10–13, 15]. Lifestyle choices, such as the use of hair dyes containing aromatic amines, have also been suggested as a potential determinant of bladder cancer, although conclusive evidence is yet to emerge [15].
Aromatic amines are known to induce DNA damage and mutations that may cause disruption of key biological pathways that may lead to cell transformation, e.g., in normal urothelium [9, 11, 16, 17]. In addition to having a genotoxic mode of action, aromatic amines, like many other chemical carcinogens, may also exert epigenetic effects of relevance to bladder carcinogenesis [6, 18–20]. Epigenetics is a fast growing field in cancer biology with tremendous potential for environmental, clinical, and translational research [21–23]. Epigenetic effects are defined as heritable changes in gene expression that do not involve alterations in the underlying DNA sequence. Aberrant DNA methylation associated with transcriptional deregulation of cancer-related genes is the best-studied epigenetic mechanism of carcinogenesis [23–28]. Aberrancies in DNA methylation patterns commonly occur in the early stages of carcinogenesis and as such, are detectable prior to clinical diagnosis of cancer. Thus, modification of DNA methylation patterns together with alterations of gene expression may serve as predictive biomarkers for early detection of cancer [29]. Furthermore, epigenetic changes intensify as cancer progresses, and are potentially reversible through pharmacological interventions or genetic manipulation [30, 31]. Therefore, epigenetic biomarkers can also be used for both prognostic and therapeutic purposes.
Investigating the epigenetic basis of bladder carcinogenesis, especially in individuals with known history of exposure to aromatic amines, e.g., specific industry workers or smokers, holds great promise for cancer biomarker discovery [6, 8, 9, 20, 32–34]. The continuous shedding of bladder cells into the urine offers a unique opportunity for non-invasive surveillance of the epigenetic landscape both before and after clinical manifestation of bladder cancer [34–36]. The non-invasively obtainable urine specimens from, e.g., occupationally exposed individuals to aromatic amines or smokers, can be analyzed over time to evaluate alterations of the epigenome during the initiation and progression of bladder cancer. Elucidating the underlying mechanisms of bladder carcinogenesis by determining the epigenetic signature of aromatic amines in a readily available surrogate tissue (i.e., urine) will be critical to developing novel diagnostic and/or prognostic biomarkers for bladder cancer.
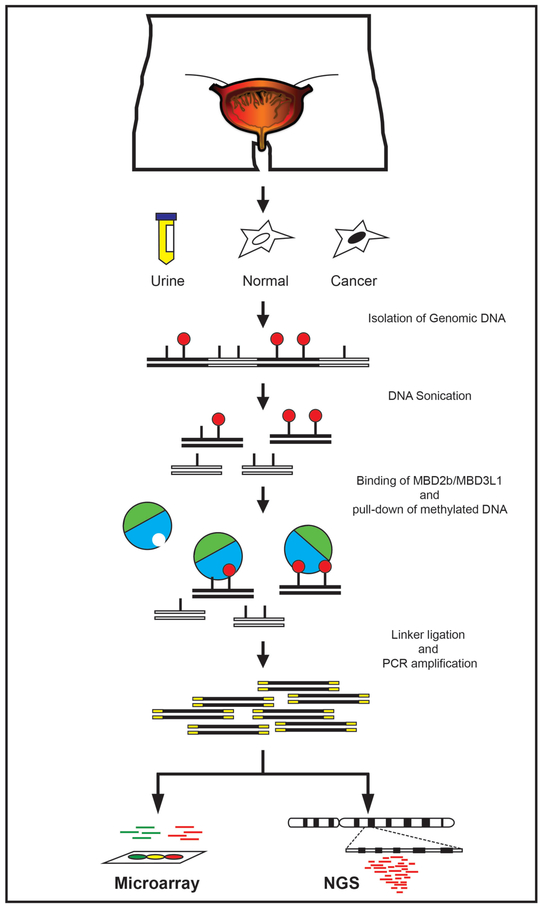
Over the past decades, an increasing number of methods have been developed to examine the DNA methylation profile at individual loci or on a genome-wide scale in a variety of experimental systems [37–40]. Here, we described a protocol for the Methylated CpG Island Recovery Assay (MIRA), a sensitive and versatile pull-down assay for enrichment of methylated DNA [41, 42]. This technique is easy to perform and allows recovery of methylated DNA without relying on the use of expensive antibodies. The MIRA (outlined in Fig. 1) is based on the ability of the methyl-binding domain (MBD) proteins, the MBD2b/MBD3L1 complex, to specifically bind methylated-CpG dinucleotides [40–42]. The MIRA-enriched DNA fractions can be used in several downstream applications, including DNA methylation analysis of single loci by real time PCR or bisulfite conversion followed by cloning and DNA sequencing [40]. MIRA is also compatible with high-throughput microarray or next-generation sequencing platforms, e.g., the Illumina Genome Analyzer (MIRA-seq). So far, several versions of MIRA-seq have been developed, all requiring ligation of specific adapters for library construction, DNA sequencing, and sequence read alignment using a reference genome [43–45]. We and others have successfully applied MIRA to detect aberrant DNA methylation in a variety of tumor types, including human melanoma, lung and breast cancer [41, 46, Tommasi, unpublished data], immortalized cell lines [47] and mice/cells treated in vivo/in vitro with prototype carcinogens [48, 49]. The MIRA technology is licensed to Active Motif (under U.S. Patent No. 7,425,415), which has developed easy-to-perform kits for MIRA (MethylCollector™ Ultra, Active Motif®) and MIRA-seq (MethylCollector™ Ultra-Seq, Active Motif ®).
Figure 1.
Outline of MIRA. This method enables analysis of the DNA methylation status in individual genes or across the entire genome. MIRA is based on the ability of the methyl-binding domain (MBD) proteins, the MBD2B/MBD3L1 complex, to specifically bind methylated CpG dinucleotides. Enriched DNA fractions can be analyzed on high-throughput microarray or NGS platforms.
2. MATERIALS
All reagents must be of molecular biology grade and solutions must be prepared using distilled milliQ water and then autoclaved/filter-sterilized.
2.1. Purification of GST-MBD2b and His-MBD3L1 proteins for MIRA
GST-tagged MBD2b and histidine-tagged MBD3L1 expression plasmids.
LB broth, LB agar, and SOC medium, for bacteriological work [50].
BL21 (DE3) Competent Cells (Agilent Technologies, Santa Clara, CA).
Isopropyl-Beta-D-Thiogalatoside (IPTG). Make a 100 mM stock solution in H2O, filter sterilize and keep at −20 oC.
Lysozyme. Make a 100 mg/ml stock solution in H2O, aliquot and keep at −20 oC.
Sodium Chloride-Tris-EDTA (STE) buffer; 150 mM NaCl, 10 mM Tris-HCl, pH 7.8, 0.1mM or 1 mM EDTA, pH 8.0.
GST-STE buffer; 150 mM NaCl, 10 mM Tris-HCl, pH 7.8, 1 mM EDTA, pH 8.0.
His-STE buffer; 150 mM NaCl, 10 mM Tris-HCl, pH 7.8, 0.1 mM EDTA, pH 8.0.
Phenylmethylsulfonylfluoride (PMSF). Make a 100 mM stock solution in isopropanol, aliquot and keep at −20 oC.
N-lauroylsarcosine. Make a 10% (w/v) stock solution in H2O, aliquot and keep at −20 oC.
Glutathione Sepharose 4B beads (GE Healthcare Life Sciences, Uppsala, Sweden).
Ni-NTA His-Bind® Resin (MilliporeSigma, Darmstadt, Germany).
GST-washing buffer; 0.1 % (v/v) Triton X-100 in phosphate buffered saline (PBS) buffer.
His-washing buffer; 50 mM NaH2PO4·H2O, 300 mM NaCl, 20 mM Imidazole. Adjust pH to 8.0 with 1 M NaOH.
GST-Elution buffer; 50 mM Tris-HCl, pH 8.5, 150 mM NaCl, 20 mM reduced glutathione, 0.1 % Triton X-100.
His-Elution buffer: 50 mM NaH2PO4·H2O, 300 mM NaCl, 250 mM Imidazole. Adjust pH to 8.0 with 1 M NaOH.
4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid) HEPES.
Protein-dialysis buffer (for both GST- and His-tagged proteins): 50 mM Hepes, pH 7.4, 150 mM NaCl, 5 mM β-mercaptoethanol, 50 % Glycerol.
Bovine Serum Albumin (BSA).
2.2. Sample preparation for MIRA
Elution buffer (EB) (Qiagen, Valencia, CA).
TE; 10 mM Tris-HCl, 1 mM EDTA, pH 8.0.
Quick-DNA™ Urine kit (Zymo Research, Irvine, CA).
2.3. MIRA enrichment and PCR amplification
Purified GST-tagged MBD2b and His-tagged MBD3L1 proteins (~1 μg each)
Sonicated DNA from JM110 bacterial strain (see Note 1).
10x MIRA binding buffer; 100 mM Tris-HCl, pH 7.9, 500 mM NaCl, 100 mM MgCl2, 10 mM DL-Dithiothreitol (DTT), 1% (v/v) Triton X-100 (see Note 2).
MagneGST Glutathione Particles and magnetic stand (Promega, Madison, WI).
Qiaquick PCR purification kit (Qiagen, Valencia, CA).
MIRA-washing buffer; 10 mM Tris-HCl, pH 7.5, 700 mM NaCl, 1 mM EDTA, 3 mM MgCl2, 0.1% (v/v) Triton X-100.
T4 DNA polymerase, 10x NEB 2 buffer, T4 DNA ligase and 10x T4 DNA ligase buffer (New England Biolabs, Ipswich, MA).
Ligation-mediated PCR (LM-PCR) Linker is obtained by annealing a long oligo: 5’ –GCGGTGACCCGGGAGATCTGAATTC-3’ with a short complementary oligo: 5’-GAATTCAGATCTCCCG-3’ (see Note 3).
Taq DNA polymerase, 10x PCR buffer, 5x Q solution (Qiagen, Valencia, CA).
Sybr Green I Nucleic Acid Gel Stain (Roche, Mannheim, Germany).
3. METHODS
3.1. Purification of GST-MBD2b and His-MBD3L1 proteins for MIRA
GST-MBD2b and His-MBD3L1 can be purified in parallel. Distinct buffers/reagents are usually needed.
3.1.1. Transformation
- In two separate tubes, transform BL21 (DE3)-competent cells with:
- GST-MBD2b protein-expressing plasmid (1 μl)
- His-MBD3L1 protein-expressing plasmid (1 μl).
Incubate tubes on ice for 30 min, at 42οC for 38 sec, and back on ice for 2 min.
Add 500 μl SOC medium and incubate at 37οC for 1 h with shaking.
- Following transformation, plate cells (100 μl) on:
- ampicillin-containing plates for GST-MBD2b
- kanamycin-containing plates for His-MBD3L1
Incubate at 37oC overnight.
3.1.2. IPTG induction
Inoculate 200 ml LB (add ampicillin for GST-MBD2b culture or kanamycin for His-MBD3L1 culture) with 30–40 well-developed bacterial colonies.
Grow in a shaker at 37oC until OD reaches 0.6 at a fixed wavelength of 600 nm.
To each flask, add 200 μl of 100 mM IPTG (0.1 mM IPTG final concentration) to induce expression of GST-tagged MBD2b or His-tagged MBD3L1 proteins.
Allow the cells to grow in the shaker at 37oC for additional 4–6 hrs.
Split cell suspension in 50 ml falcon tubes (4x) and centrifuge at 3500 xg for 15 min at 4oC. Discard supernatant.
Cell pellets can be processed immediately or kept at −80oC for several months.
3.1.3. Protein purification
- Resuspend the cell pellet (from a single falcon tube) in 10 ml of:
- GST-STE buffer containing 100 μg/ml lysozyme for GST-MBD2b;
- His-STE buffer containing 100 μg/ml lysozyme for His-MBD3L1.
To each tube, add 100 μl of 100 mM PMSF (to a final concentration of 1 mM PMSF) and incubate on ice for 10 min.
Lyse bacterial cells by adding 1 ml of 10% N-lauroylsarcosine.
Sonicate bacterial lysate until it clears up and loses viscosity.
Add 1 ml of 10% Triton-X to the lysate and vortex it for 20 sec.
Centrifuge lysate at 3500 xg for 15 min (4oC) and transfer supernatant into a new tube (~12 ml).
- To the cleared lysate add:
- 0.1 ml 50% slurry Glutathione Sepharose 4B beads for GST-MBD2b;
- 0.1 ml Ni-NTA Agarose beads for His-MBD3L1.
Mix gently by shaking at 4oC for 30–45 min.
Pellet the beads at 1000 xg for 1 min.
- Wash with:
- 10 ml of GST-washing buffer for GST-MBD2b;
- 10 ml of His-washing buffer for His-MBD3L1.
Invert tubes several times, then collect beads by centrifugation at 1000 xg for 1 min.
Repeat washes two more times.
3.1.4. Elution and dialysis
- Elute proteins from beads with:
- 1 ml GST-Elution buffer for ~4 hrs at 4oC on a rotating platform for GST-MBD2b;
- 1 ml His-Elution buffer for 30 min at 4oC on a rotating platform for His-MBD3L1.
- Dialyze the eluted proteins against:
- 1–2 liters of 1x PBS (+PMSF) for 5 hrs at 4oC, and then against 1–2 liters of protein-dialysis buffer (+PMSF) overnight at 4oC.
- After dialysis, check protein integrity and concentration on a 10% SDS-PAGE gel using BSA as a standard.
Aliquots of purified MBD2b and MBD3L1 proteins can be kept at −20oC for several months.
3.2. Sample preparation for MIRA
3.2.1. DNA isolation
High molecular weight genomic DNA can be isolated from cells and tissues using standard phenol/chloroform extraction protocols [50]. Alternatively, commercially available kits, such as the DNeasy Blood & Tissue Kit (Qiagen, Valencia, CA) can be used.
Genomic DNA can be extracted from urine specimens with the Quick-DNA™ Urine kit (Zymo Research, Irvine, CA).
3.2.2. Fragmentation of genomic DNA
We routinely use a Bioruptor® sonicator (Diagenode, Denville, NJ) to shear genomic DNA to generate 200- to 600-bp fragments (see Note 4).
Resuspend 500–700 ng genomic DNA to a final volume of 200 μl (with EB or TE buffer).
Sonication is performed in apposite tubes using the medium power setting with alternating 30 sec on/30 sec off intervals for a total of 15 min. Avoid overheating.
Check an aliquot on 2% agarose gel.
Keep 10–20 ng sonicated DNA to use as input (non-MIRA enriched fraction).
3.3. MIRA
3.3.1. Binding of MBD proteins to fragmented DNA
In a 1.5 ml Eppendorf tube, mix: 1x NEB buffer 2, 0.1% Triton X-100, 0.5 μg sonicated JM110 DNA, 1 μg purified GST-MBD2b, 1 μg purified His-MBD3L1, and H2O to a final volume of 100 μl.
Mix by pipetting and pre-incubate at 4oC for 20 min on a rotating platform. Rotate at a low speed to prevent foaming. This step is required for MBD2b/MBD3L1 complex formation.
Add 500–700 ng sonicated genomic DNA to the 100 μl binding mix. Adjust final volume to 400 μl (with EB buffer).
Incubate at 4oC overnight (or for at least 5 hrs) on a rotating platform.
3.3.2. Washing and pre-blocking of Magne GST beads
Add 2.5 μl of MagneGST Glutathione Particles into 1 ml of 1x PBS containing 0.1 % Triton X-100 and invert tubes several times.
Capture magnetic beads using a magnetic stand and carefully decant the supernatant.
Add 1 ml of 1x PBS / 0.1% Triton X-100 into each tube.
Repeat washes for 2–3 times.
To decrease non-specific background, pre-block the washed beads with a solution of: 1x NEB buffer 2, 0.1% Triton X-100, 0.5 μg sonicated JM110 DNA, and H2O up to 400 μl.
Mix by pipetting and incubate at 4oC for 20 min on a rotating platform.
Remove supernatant with the aid of a magnetic stand.
Add 400 μl of protein-DNA mix (from 3.3.1.) to the pre-blocked beads.
Incubate at 4oC for ~2 hrs on a rotating platform.
3.3.3. Recovery of the MIRA-enriched DNA fraction
Capture the beads with the aid of a magnetic stand and carefully remove the supernatant.
Add 800 μl of MIRA-washing buffer into the tube and invert 4–5 times. Capture the beads on a magnetic stand. Discard the supernatant.
Wash beads (carrying the methylated CpG fraction) for 2–3 times with 1 ml of MIRA-washing buffer. Following beads pulldown, discard supernatant.
Purify the CpG-enriched fraction from the beads with the Qiaquick PCR purification kit according to the manufacturer’s instructions.
Elute the CpG-rich fraction from the column with 50 μl of EB buffer.
3.3.4. Generation of blunt-ended DNA
Following sonication, DNA fragments must undergo an end-treatment filling step for successful ligation of the blunt-ended linker adaptor. *Remember to include the input sample (non-MIRA enriched DNA fraction) in parallel at this point.
To each input and MIRA-enriched DNA sample (50 μl) add: 1x NEB buffer 2, 100 μM dNTPs, 1x BSA, 0.6 U T4 DNA polymerase, and H2O up to a final volume of 60 μl.
Mix by pipetting and incubate at 12oC for 20 min.
Add 300 μl PBI buffer to each tube and purify according to the Qiaquick protocol (Qiagen).
Elute in 50 μl EB buffer.
Speed vac to 5 μl.
3.3.5. Linker ligation and PCR amplification
For linker ligation, to each blunt-ended DNA sample (5 μl), add 5 μl ligation mix containing: 15 μM double-stranded LM-PCR linker, 1x T4 DNA ligase buffer, and 400 U T4 DNA ligase.
Incubate at 16oC overnight.
For PCR amplification, add to each tube: 1x PCR Buffer, 1x Q solution, 1.2 mM MgCl2, 0.35 mM dNTP, 0.375x Sybr Green, 5 U Taq Polymerase, and H2O up to 100 μl.
Incubate samples in a real-time PCR machine using the following cycling conditions: 72oC for 15 min; 95oC for 3 min; [95oC for 30 sec, 60oC 20 sec] for 14–16 cycles; 72oC for 3 min and 30 sec (see Note 5).
Following PCR amplification, purify DNA by Qiaquick purification kit (Qiagen) and elute in 50 μl EB buffer.
Measure the concentration by a spectrophotometer or Nanodrop.
At this point, input and MIRA-enriched DNA from experimental and control samples can be labeled and examined by microarray analysis. Alternatively, samples can be ligated to specific adaptors followed by library construction and analysis on NGS platforms (Fig. 1).
4. NOTES
JM110 is a bacterial strain that lacks both DNA adenine methylation (dam) and DNA cytosine methylation (dcm) activities. Purified JM110 DNA is sonicated to an average length of 400–500 bp. Sonicated JM110 DNA is used in the binding reaction to decrease non-specific binding of DNA to MBD proteins and/or beads.
10x NEB 2 buffer (New England Biolabs, Ipswich, MA) can be used as an alternative buffer. Remember to add Triton X-100 to a final concentration of 0.1%.
The LM-PCR linker is prepared by combining 50 μl of 100 μM long oligo with 50 μl of 100 μM short oligo. The mix (50 μM) is incubated for 3 min in a boiling water bath, and allowed to slowly cool down to room temperature. The annealed double-stranded linker is kept in small aliquots at −20°C.
Alternatively, genomic DNA can be restriction digested with MseI (5’-TTAA-3’), which cuts outside the CpG islands. Different sets of oligos are required to make the linker adaptor. Sonication is preferable to MseI digestion because it does not introduce significant sequence bias.
The initial step at 72°C is required to fill in the 3’ ends of the double-stranded linker adaptor. PCR cycling is monitored in a real-time thermocycler and reactions are stopped immediately before reaching the amplification plateau (14–16 cycles are usually sufficient). PCR reactions can be scaled down to 10–50 μl (final volume).
5. ACKNOWLEDGEMENTS
ST and AB declare no conflicts of interest.
Work of the authors is funded by grants from the National Institute of Dental and Craniofacial Research of the National Institutes of Health (1R01DE026043–01) to AB and from the University of California Tobacco-Related Disease Research Program (TRDRP-25IP-0001) to ST.
6. REFERENCES
- 1.Stewart BW, Wild CP (eds) (2014) World Cancer Report 2014. International Agency for Research on Cancer (IARC), Lyon, France [Google Scholar]
- 2.Antoni S, Ferlay J, Soerjomataram I, Znaor A, Jemal A, Bray F (2016) Bladder Cancer Incidence and Mortality: A Global Overview and Recent Trends. Eur Urol doi: 10.1016/j.eururo.2016.06.010 [DOI] [PubMed] [Google Scholar]
- 3.Kamat AM, Hahn NM, Efstathiou JA, Lerner SP, Malmstrom PU, Choi W, Guo CC, Lotan Y, Kassouf W (2016) Bladder cancer. Lancet. doi: 10.1016/S0140-6736(16)30512-8 [DOI] [PubMed] [Google Scholar]
- 4.Siegel RL, Miller KD, Jemal A (2016) Cancer statistics, 2016. CA Cancer J Clin 66 (1):7–30 [DOI] [PubMed] [Google Scholar]
- 5.Carradori S, Cristini C, Secci D, Gulia C, Gentile V, Di Pierro GB (2012) Current and emerging strategies in bladder cancer. Anticancer Agents Med Chem 12 (6):589–603 [DOI] [PubMed] [Google Scholar]
- 6.Besaratinia A, Cockburn M, Tommasi S (2013) Alterations of DNA methylome in human bladder cancer. Epigenetics 8 (10):1013–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Griffiths TR, on behalf of Action on Bladder Cancer (2013) Current perspectives in bladder cancer management. Int J Clin Pract 67 (5):435–448 [DOI] [PubMed] [Google Scholar]
- 8.Miremami J, Kyprianou N (2014) The promise of novel molecular markers in bladder cancer. Int J Mol Sci 15 (12):23897–23908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Netto GJ (2012) Molecular biomarkers in urothelial carcinoma of the bladder: are we there yet? Nat Rev Urol 9 (1):41–51 [DOI] [PubMed] [Google Scholar]
- 10.Boffetta P (2008) Tobacco smoking and risk of bladder cancer. Scand J Urol Nephrol Suppl (218):45–54 [DOI] [PubMed] [Google Scholar]
- 11.Besaratinia A, Tommasi S (2013) Genotoxicity of tobacco smoke-derived aromatic amines and bladder cancer: current state of knowledge and future research directions. FASEB J 27 (6):2090–2100 [DOI] [PubMed] [Google Scholar]
- 12.Kiriluk KJ, Prasad SM, Patel AR, Steinberg GD, Smith ND (2012) Bladder cancer risk from occupational and environmental exposures. Urol Oncol 30 (2):199–211 [DOI] [PubMed] [Google Scholar]
- 13.U.S. Department of Health and Human Services (2014) The health consequences of smoking - 50 years of progress. A report of the Surgeon General. U.S. Department of Health and Human Services. Public Health Service. Office of the Surgeon General; Rockville, MD [Google Scholar]
- 14.Freedman ND, Silverman DT, Hollenbeck AR, Schatzkin A, Abnet CC (2011) Association between smoking and risk of bladder cancer among men and women. JAMA 306 (7):737–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Internationa Agency for Research on Cancer (IARC) (2010) Some Aromatic Amines, Organic Dyes, and Related Exposures. In: IARC Monographs on the Evaluation of Carcinogenic Risk to Humans, vol 99 Lyon, France: [PMC free article] [PubMed] [Google Scholar]
- 16.Scelo G, Brennan P (2007) The epidemiology of bladder and kidney cancer. Nat Clin Pract Urol 4 (4):205–217 [DOI] [PubMed] [Google Scholar]
- 17.Stern MC, Lin J, Figueroa JD, Kelsey KT, Kiltie AE, Yuan JM, Matullo G, Fletcher T, Benhamou S, Taylor JA, Placidi D, Zhang ZF, Steineck G, Rothman N, Kogevinas M, Silverman D, Malats N, Chanock S, Wu X, Karagas MR, Andrew AS, Nelson HH, Bishop DT, Sak SC, Choudhury A, Barrett JH, Elliot F, Corral R, Joshi AD, Gago-Dominguez M, Cortessis VK, Xiang YB, Gao YT, Vineis P, Sacerdote C, Guarrera S, Polidoro S, Allione A, Gurzau E, Koppova K, Kumar R, Rudnai P, Porru S, Carta A, Campagna M, Arici C, Park SS, Garcia-Closas M, International Consortium of Bladder C (2009) Polymorphisms in DNA repair genes, smoking, and bladder cancer risk: findings from the international consortium of bladder cancer. Cancer Res 69 (17):6857–6864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jirtle RL, Skinner MK (2007) Environmental epigenomics and disease susceptibility. Nat Rev Genet 8 (4):253–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cortessis VK, Thomas DC, Levine AJ, Breton CV, Mack TM, Siegmund KD, Haile RW, Laird PW (2012) Environmental epigenetics: prospects for studying epigenetic mediation of exposure-response relationships. Hum Genet 131 (10):1565–1589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schulz WA, Goering W (2016) DNA methylation in urothelial carcinoma. Epigenomics. doi: 10.2217/epi-2016-0064 [DOI] [PubMed] [Google Scholar]
- 21.Sandoval J, Esteller M (2012) Cancer epigenomics: beyond genomics. Curr Opin Genet Dev 22 (1):50–55 [DOI] [PubMed] [Google Scholar]
- 22.You JS, Jones PA (2012) Cancer genetics and epigenetics: two sides of the same coin? Cancer Cell 22 (1):9–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Feinberg AP, Koldobskiy MA, Gondor A (2016) Epigenetic modulators, modifiers and mediators in cancer aetiology and progression. Nat Rev Genet 17 (5):284–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Suzuki MM, Bird A (2008) DNA methylation landscapes: provocative insights from epigenomics. Nat Rev Genet 9 (6):465–476 [DOI] [PubMed] [Google Scholar]
- 25.Kulis M, Esteller M (2010) DNA methylation and cancer. Adv Genet 70:27–56 [DOI] [PubMed] [Google Scholar]
- 26.Jones PA (2012) Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet 13 (7):484–492 [DOI] [PubMed] [Google Scholar]
- 27.Besaratinia A, Tommasi S (2014) Epigenetics of human melanoma: promises and challenges. J Mol Cell Biol 6 (5):356–367 [DOI] [PubMed] [Google Scholar]
- 28.Beekman R, Kulis M, Martín-Subero JI (2016) The DNA Methylomes of Cancer In: Fraga M, Fernandez AF (eds) Epigenomics in Health and Disease. Elsevier Inc., pp 183–207 [Google Scholar]
- 29.Verma M (2015) The Role of Epigenomics in the Study of Cancer Biomarkers and in the Development of Diagnostic Tools. Adv Exp Med Biol 867:59–80 [DOI] [PubMed] [Google Scholar]
- 30.Rodriguez-Paredes M, Esteller M (2011) Cancer epigenetics reaches mainstream oncology. Nat Med 17 (3):330–339 [DOI] [PubMed] [Google Scholar]
- 31.Costa-Pinheiro P, Montezuma D, Henrique R, Jeronimo C (2015) Diagnostic and prognostic epigenetic biomarkers in cancer. Epigenomics 7 (6):1003–1015 [DOI] [PubMed] [Google Scholar]
- 32.Chihara Y, Kanai Y, Fujimoto H, Sugano K, Kawashima K, Liang G, Jones PA, Fujimoto K, Kuniyasu H, Hirao Y (2013) Diagnostic markers of urothelial cancer based on DNA methylation analysis. BMC Cancer 13:275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harb-de la Rosa A, Acker M, Kumar RA, Manoharan M (2015) Epigenetics application in the diagnosis and treatment of bladder cancer. Can J Urol 22 (5):7947–7951 [PubMed] [Google Scholar]
- 34.Wu P, Cao Z, Wu S (2016) New Progress of Epigenetic Biomarkers in Urological Cancer. Dis Markers 2016:9864047. doi: 10.1155/2016/9864047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chung W, Bondaruk J, Jelinek J, Lotan Y, Liang S, Czerniak B, Issa JP (2011) Detection of bladder cancer using novel DNA methylation biomarkers in urine sediments. Cancer Epidemiol Biomarkers Prev 20 (7):1483–1491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sapre N, Anderson PD, Costello AJ, Hovens CM, Corcoran NM (2014) Gene-based urinary biomarkers for bladder cancer: an unfulfilled promise? Urol Oncol 32 (1):48 e49–17 [DOI] [PubMed] [Google Scholar]
- 37.Laird PW (2010) Principles and challenges of genomewide DNA methylation analysis. Nat Rev Genet 11 (3):191–203 [DOI] [PubMed] [Google Scholar]
- 38.Fouse SD, Nagarajan RO, Costello JF (2010) Genome-scale DNA methylation analysis. Epigenomics 2 (1):105–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Taylor KH, Shi H, Caldwell CW (2010) Next generation sequencing: advances in characterizing the methylome. Genes 1 (2):143–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Olkhov-Mitsel E, Bapat B (2012) Strategies for discovery and validation of methylated and hydroxymethylated DNA biomarkers. Cancer Med 1 (2):237–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tommasi S, Karm DL, Wu X, Yen Y, Pfeifer GP (2009) Methylation of homeobox genes is a frequent and early epigenetic event in breast cancer. Breast Cancer Res 11 (1):R14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mitchell N, Deangelis JT, Tollefsbol TO (2011) Methylated-CpG Island Recovery Assay. Methods Mol Biol 791:125–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Choi JH, Li Y, Guo J, Pei L, Rauch TA, Kramer RS, Macmil SL, Wiley GB, Bennett LB, Schnabel JL, Taylor KH, Kim S, Xu D, Sreekumar A, Pfeifer GP, Roe BA, Caldwell CW, Bhalla KN, Shi H (2010) Genome-wide DNA methylation maps in follicular lymphoma cells determined by methylation-enriched bisulfite sequencing. PloS One 5 (9): e13020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Almamun M, Levinson BT, Gater ST, Schnabel RD, Arthur GL, Davis JW, Taylor KH (2014) Genome-wide DNA methylation analysis in precursor B-cells. Epigenetics 9 (12):1588–1595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Green BB, McKay SD, Kerr DE (2015) Age dependent changes in the LPS induced transcriptome of bovine dermal fibroblasts occurs without major changes in the methylome. BMC Genomics 16:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tommasi S, Kim SI, Zhong X, Wu X, Pfeifer GP, Besaratinia A (2010) Investigating the epigenetic effects of a prototype smoke-derived carcinogen in human cells. PloS One 5 (5):e10594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tommasi S, Zheng A, Weninger A, Bates SE, Li XA, Wu X, Hollstein M, Besaratinia A (2013) Mammalian cells acquire epigenetic hallmarks of human cancer during immortalization. Nucleic Acids Res 41 (1):182–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tommasi S, Zheng A, Yoon JI, Li AX, Wu X, Besaratinia A (2012) Whole DNA methylome profiling in mice exposed to secondhand smoke. Epigenetics 7 (11):1302–1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tommasi S, Zheng A, Yoon JI, Besaratinia A (2014) Epigenetic targeting of the Nanog pathway and signaling networks during chemical carcinogenesis. Carcinogenesis 35 (8):1726–1736 [DOI] [PubMed] [Google Scholar]
- 50.Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]