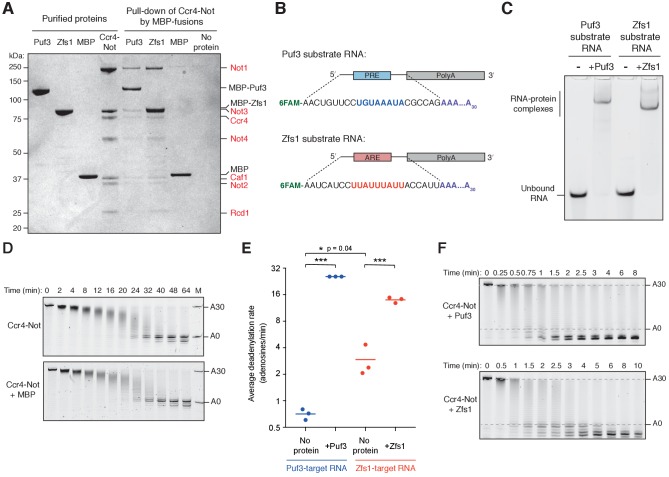
(
A) Coomassie-stained SDS-PAGE of pull-down assay showing binding of Ccr4-Not (subunits labelled in red) to immobilized MBP-Puf3 and MBP-Zfs1. (
B) Schematic diagram of RNA substrates showing 5ʹ 6-FAM fluorescent label, position of Pumilio-response element (PRE; blue), AU-rich element (ARE; red), and 30-adenosine poly(A) tail. (
C) Electrophoretic mobility shift assay (EMSA) showing the protein-RNA complexes used as substrates for deadenylation assays. Puf3 or Zfs1 (250 nM) was incubated with labelled RNA (200 nM) for 15 min at room temperature before resolving on a native polyacrylamide gel. (
D) Control deadenylation assay with Puf3-target RNA showing that MBP alone does not have an effect on the deadenylation activity of Ccr4-Not. M is marker for RNA with and without a poly(A)
30 tail. (
E) Average rates of Ccr4-Not-mediated deadenylation in the presence of Puf3 or Zfs1. Reaction rates were calculated by densitometric analysis of denaturing polyacrylamide gels. The most abundant RNA size decreased linearly with time, and average reaction rates in number of adenosines removed/min were determined by linear regression on triplicate measurements. Each experiment is presented as a single data point, and the mean of each triplicate experiment is plotted as a horizontal line. Statistical significance was calculated by one-way ANOVA in GraphPad Prism. *p=0.04; ***p=0.001. (
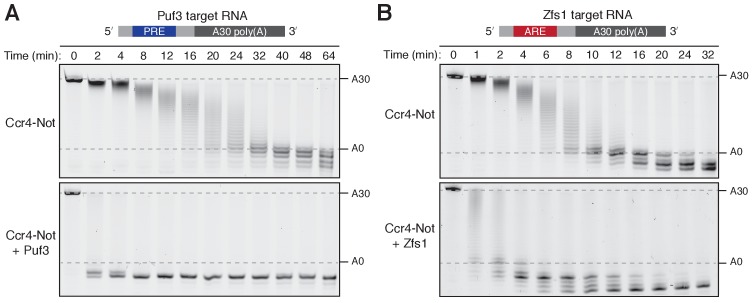
F) Fully-deadenylated and non-deadenylated RNA exist simultaneously in reactions with Ccr4-Not and Puf3 or Zfs1. Experiments were performed as in
Figure 1 with shorter time increments.