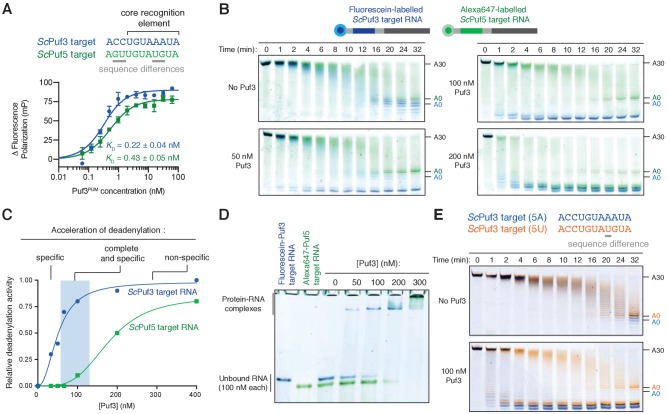
Figure 4. Puf3 distinguishes between similar RNA motifs.
(A) Fluorescence polarization experiments assaying the binding of Puf3 RNA-binding domain to RNA containing a ScPuf3 motif (blue) or a ScPuf5 motif (green). Error bars represent the standard error in triplicate measurements. (B) Deadenylation of a mixture of fluorescein-labelled ScPuf3-target RNA (blue) and Cy3-labelled ScPuf5-target RNA (green) analyzed by denaturing PAGE. Gels were scanned to detect each fluorophore before signals were overlaid. (C) Selective deadenylation occurs within a narrow range of Puf3 concentrations. Plot of relative deadenylation rates of ScPuf3-target RNA and ScPuf5-target RNA in reactions as in (B) at a series of Puf3 concentrations. Reactions were quantified based on the fraction of RNA fully deadenylated within 8 min. (D) Electrophoretic mobility shift assay showing the binding of Puf3 to a mixture of fluorescein-labelled ScPuf3-target RNA (blue) and Cy3-labelled ScPuf5-target RNA (green). The concentration of each RNA was 100 nM. (E) Deadenylation of a mixture of fluorescein-labelled ScPuf3(5A)-target RNA (blue) and Alexa647-labelled ScPuf3(5U)-target RNA (orange) analyzed by denaturing PAGE. Gels were scanned to detect each fluorophore before signals were overlaid.