Figure 1.
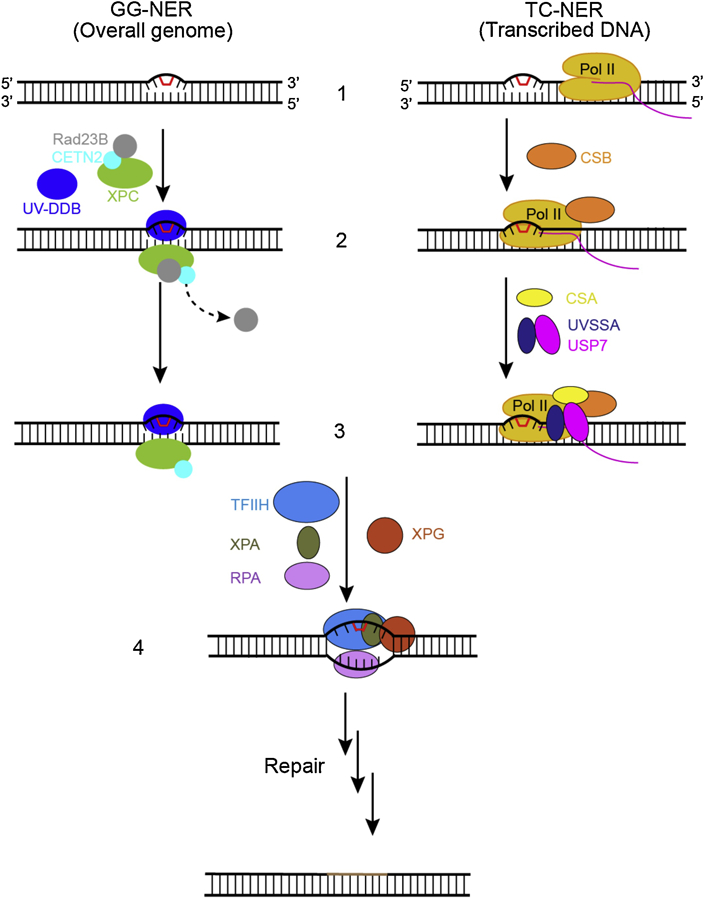
The recognition steps of two subpathways of nucleotide excision repair. In the global genome nucleotide excision repair subpathway (GG-NER), left, the damage sensor XPC, in complex with UV excision repair protein RAD23 homologue B (RAD23B) and centrin 2 (CETN2), which binds the non-damaged strand opposite the lesion with the help of the UV– DDB complex (step 2, left). Binding of the XPC complex to the damaged site results in RAD23B dissociation from the complex (step 3, left). In the transcription-coupled subpathway (TC-NER), damage recognition is initiated by the stalling of RNA polymerase II (Pol II). The stalled Pol II recruits CSB (step 2, right) and the Pol II–CSB complex serves as a platform to further recruitment of downstream repair factors such as CSA and UVSSA-USP7 (step 3, right). After damage recognition (step1–3), the TFIIH complex is recruited to the lesion in both GG-NER and TC-NER, along with XPA, RPA, and XPG (step 4). Once DNA lesion is verified, the damage-containing DNA short fragment is removed via dual incisions, new DNA fragment is synthesized, and NER reaction is completed through sealing the final nick by DNA ligase.
