Figure 2.
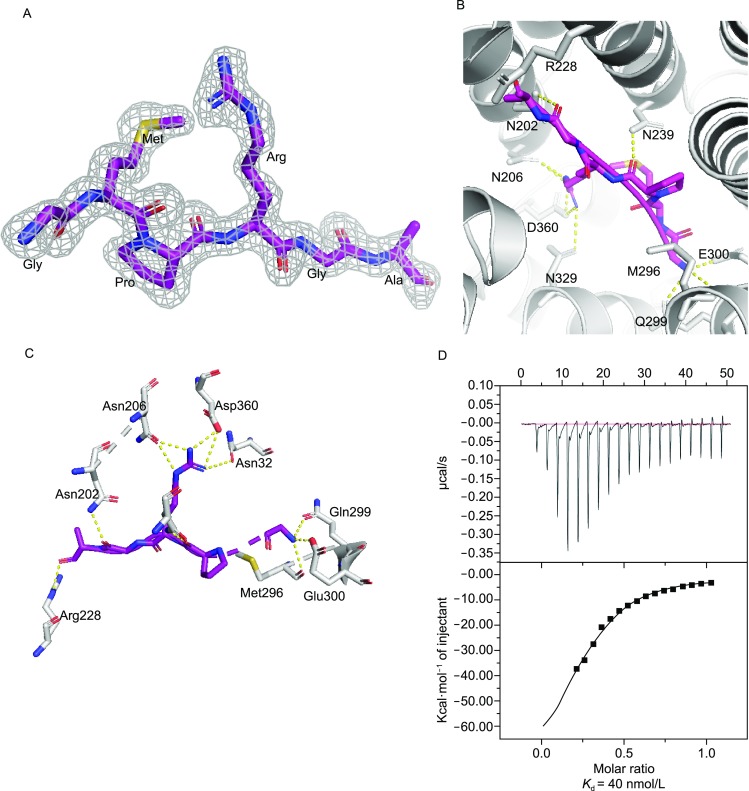
Recognition of AimP hexapeptide (GMPRGA) by AimR from SPbeta phage. (A) Electron density map of the AimP bound to AimR. The 2Fo-Fc map, calculated by simulated annealing without peptide in the structure, is shown contoured at 1.5 σ as a gray grid with the peptide in sticks colored by atom type. (B) The binding mode between AimR and AimP. AimR is shown as cartoon colored in white, whereas the residues that interact with AimP (N202, N206, N239, Q299, E300, N329 and D360) are shown as sticks colored by atom type. AimP is shown in stick representation with purple carbons. Hydrogen bonds are depicted as dotted yellow lines. (C) Similar to (B), but showing only interacting residues from another angle of view. (D) Binding affinity of wild-type AimR for AimP measured by ITC. The concentration of AimP used in assay was 200–500 μmol/L, and the concentration of AimR was 10–20 μmol/L
