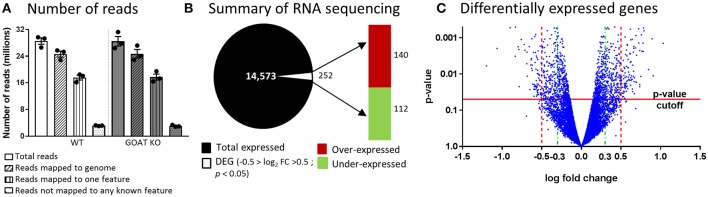
Figure 2.
RNA sequencing analysis of ovaries from juvenile GOAT KO versus WT mice. (A) Summary of number of reads and their mapping to genome. (B) Summary of RNA sequencing. Total number of identified transcripts are presented as black and differentially expressed genes (DEGs; −0.5 > logFC > 0.5; p < 0.05) are presented as white. The red bar represents the number of over-expressed transcripts and the green bar represents the number of under-expressed transcripts in the ovaries of juvenile GOAT KO mice. (C) Volcano plot for differentially expressed genes, where the p-value (Y axis) is plotted against the log fold change (X axis). p-value cutoff of 0.05 is represented by horizontal red line. Log fold change filters are represented by vertical dashed lines. Red dashed lines represent a filter set to ±0.5 that was used to test for over-representation of Gene Ontology terms and for pathway enrichment analysis. Green dashed lines represent a filter set to ±0.3, for the analysis using the Ingenuity Pathway Analysis (IPA) platform.