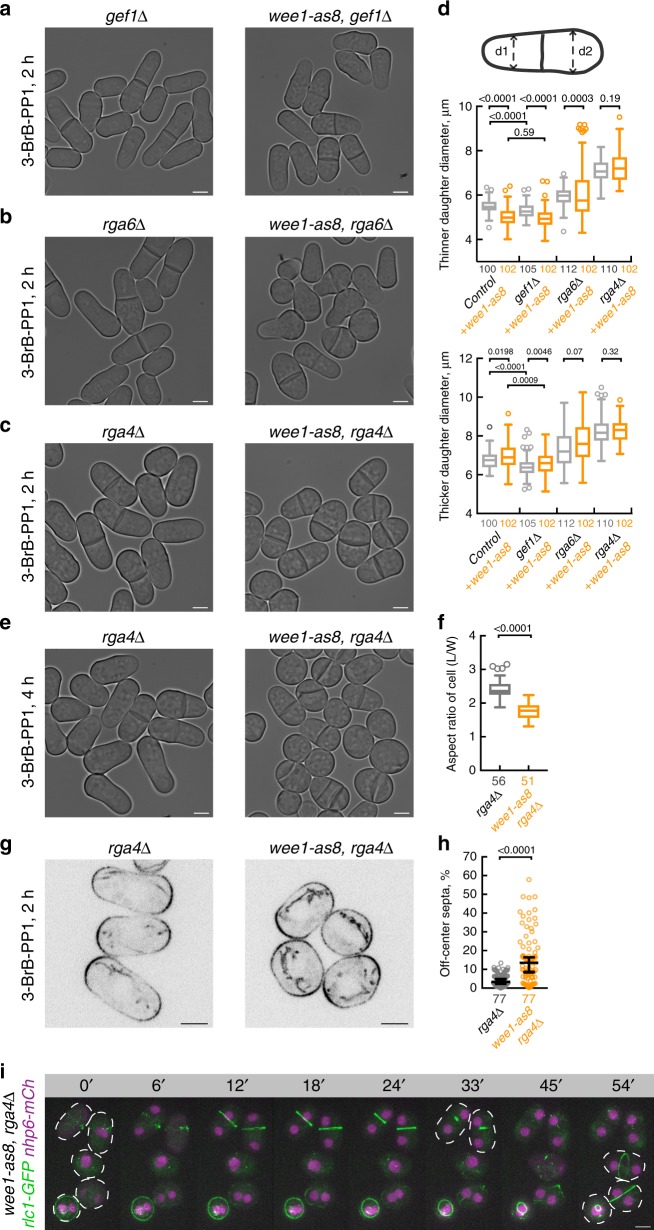
Fig. 3.
Rga4-dependent rescaling of cellular geometry is essential for medial division plane positioning. a–c Single z-plane bright-field micrographs of S. japonicus cells of indicated genotypes incubated with 20 μM 3-BrB-PP1 (right) for 2 h. Note that Rga4 deficiency prevents hyperpolarization of growth upon Wee1 inhibition. d Quantifications of cell diameters in the “thinner” (top) and “thicker” daughter (bottom) cell at cytokinesis, when 3-BrB-PP1 treatment is combined with genetic disruption of Cdc42 module regulators shown in (a–c). e wee1-as8 rga4Δ cells incubated with 20 μM 3-BrB-PP1 (right) for 4 h show severe division site positioning defects. Shown are bright-field micrographs. f Quantifications of cellular aspect ratio at division of cells shown in (e). g Single z-plane spinning-disk confocal micrographs of Pom1-GFP-expressing rga4Δ and wee1-as8 rga4Δ cells treated with 20 μM 3-BrB-PP1 for 2 h. h A plot summarizing the accuracy of division plane positioning in cells shown in (e). Deviation from the geometric center of the cell is indicated on y-axis. Black bars represent sample median. i Time-lapse montage of maximum intensity z-projected spinning-disk confocal micrographs of 3-BrB-PP1-treated wee1-as8 rga4Δ S. japonicus cells expressing Rlc1-GFP and the nucleoplasmic protein Nhp6-mCherry. Cell boundaries are outlined by white dashed lines. Wee1-inhibited S. japonicus cells lacking Rga4 fail to anchor the actomyosin ring medially following the semi-open mitosis, as indicated by the dispersal and nuclear re-import of Nhp6. a–c, e, g, i Scale bars represent 5 μm. d, f, h n indicated in figures; p values derived using Kolmogorov–Smirnov test