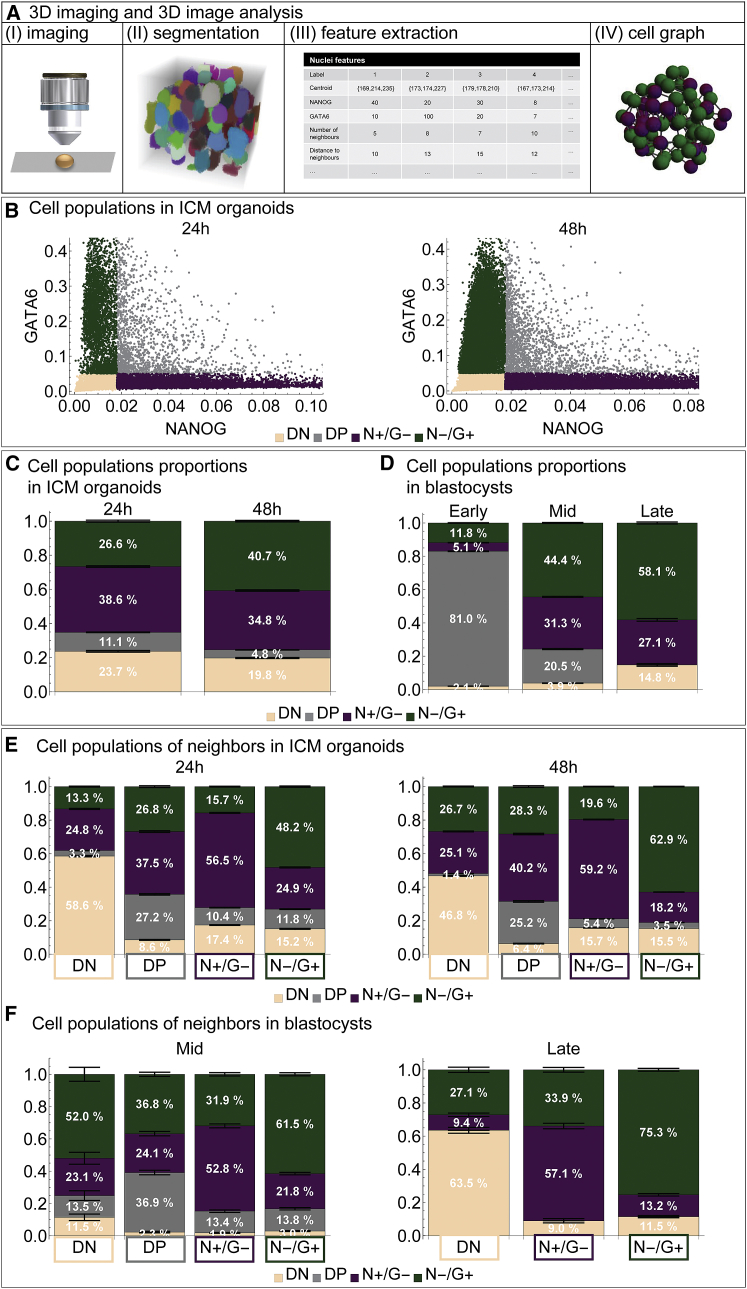
Figure 7.
Lineage composition and spatial distribution of GATA6 and NANOG in ICM organoids resemble those of mid- and late mouse blastocysts. (A) Three-dimensional imaging and three-dimensional image analysis form the basis for a quantitative comparison between ICM organoids of mouse tet::GATA4 ES cells and blastocysts. Quantitative data of early, mid-, and late blastocysts were taken from Saiz et al. (22) (B) Fluorescence intensity levels of GATA6 and NANOG of individual cells in ICM organoids after 24 and 48 h. The data points are colored by cell population. (C and D) Lineage composition is shown as percentage of the total number of cells in ICM organoids after 24 and 48 h and in the ICM of early, mid-, and late blastocysts. (E and F) Lineage composition of neighboring cells is shown as percentage of the total of neighboring cells in ICM organoids after 24 and 48 h and the ICM of mid- and late blastocysts. The error bars indicate the standard error of the mean. The number of independent experiments for ICM organoids or blastocysts is, respectively, 76, 147. DN: double negative (NANOG−/GATA6−), DP: double positive (NANOG+/GATA6+), N+/G− (NANOG+/GATA6−), N−/G+ (NANOG−/GATA6+). To see this figure in color, go online.