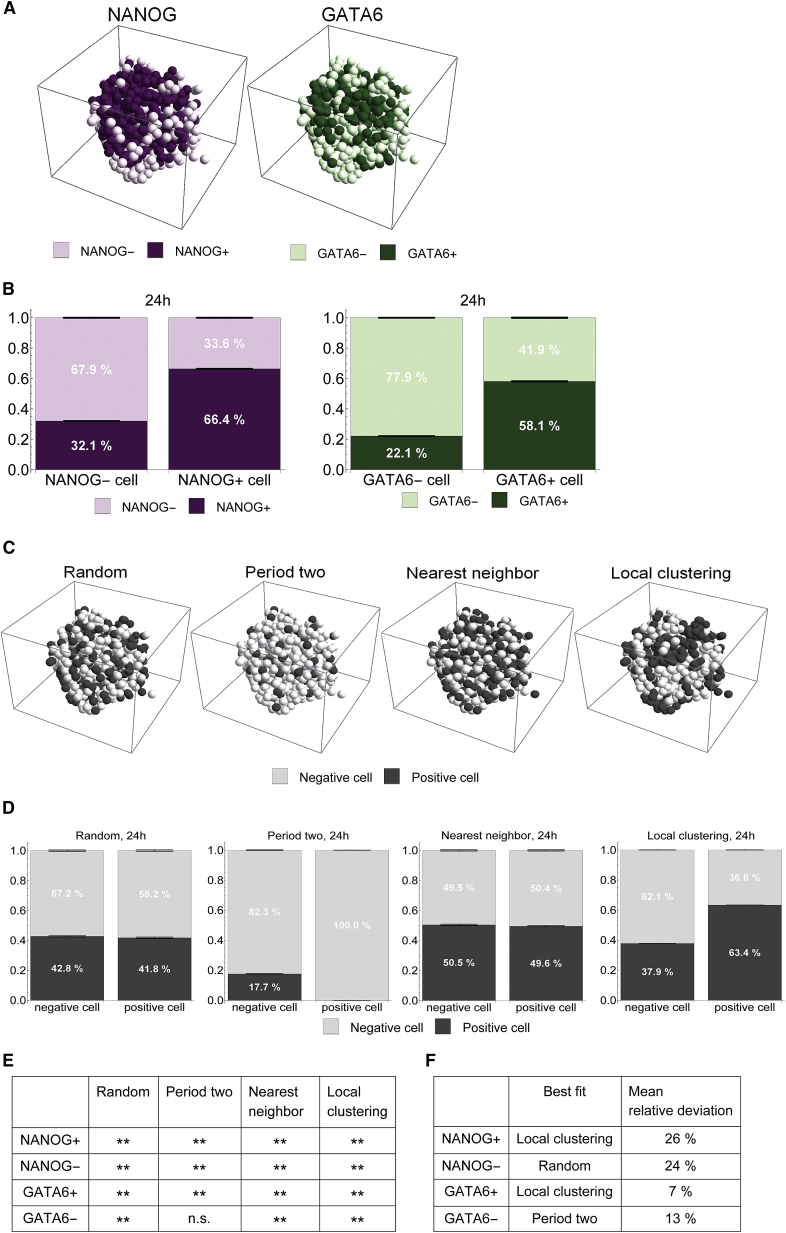
Figure 8.
Mutual exclusive pattern in 1-day-old ICM organoids exhibits local cell fate clustering. (A) Spatial distribution of NANOG-positive and negative cells or GATA6-positive and negative cells, respectively, in a 1-day-old ICM organoid (positive: N+/G− or G+/N−, respectively, and DP; negative: N−/G+ or G−/N+, respectively and DN) (B) Lineage composition of neighboring cells is shown as percentage of the total number of neighboring cells in ICM organoids after 24 h. The error bars indicate the standard error of the mean. (C) Simulation of three artificial patterns describing the salt-and-pepper patterning (for more information, please see Materials and Methods). A negative cell is NANOG-negative or GATA6-negative, respectively. A positive cell is NANOG-positive or GATA6-positive, respectively. (D) Lineage composition of neighboring cells is shown as percentage of the total of neighboring cells in the three simulated artificial patterns. The error bars indicate the standard error of the mean. (E) Statistical comparison of the data from 24-h-old ICM organoids with the three artificial patterns (∗∗p < 0.01, n.s.: not significant; for details, Wilcoxon-Mann-Whitney test with Bonferroni correction). (F) The closest fit of the simulated pattern to the experimental data is expressed as mean relative deviation, i.e., , where is the mean of the simulated data and the mean of the experimental data. Number of independent experiments for ICM organoids: 34. To see this figure in color, go online.