Abstract
GM2-synthase produces sialic acid–containing glycosphingolipids called gangliosides, and its mRNA overexpression and the gangliosides it generates are linked to tumor progression, migration, and suppression of tumor-specific host immune responses. However, the mechanism underlying GM2-synthase de-repression in renal cell carcinoma (RCC) is poorly understood. Here, we demonstrate that higher GM2-synthase mRNA expression levels in various cancer cells and in human RCC tumors correlate with higher histone acetylation levels (H3K9, H3K14, or both) at region +38/+187 relative to the transcription start site (TSS) of the GM2-synthase gene than in normal kidney epithelial (NKE) cells or healthy adjacent tissues. An increase in GM2-synthase mRNA expression in cells treated with a histone deacetylase (HDAC) inhibitor was accompanied by increased histone acetylation levels at this promoter region. DNA methylation around the TSS was absent in both RCC cell lines and NKE cells. Of note, both the transcription factor Sp1 and corepressor HDAC1 associated with the +38/+187 region when the GM2-synthase gene was repressed in NKE and tumor-adjacent tissues, indicating plausible site-specific repressive roles of HDAC1 and Sp1 in GM2-synthase mRNA expression. Site-directed mutagenesis of the Sp1-binding site within the +38/+187 region relieved repressed luciferase activity of this region by limiting HDAC1 recruitment. Moreover, Sp1 or HDAC1 knock down increased GM2-synthase transcription, and butyrate-mediated activation of GM2-synthase mRNA expression in SK-RC-45 cells was accompanied by Sp1 and HDAC1 loss from the +38/+187 region. Taken together, we have identified an epigenetic mechanism for the de-repression of the GM2-synthase gene in RCC.
Keywords: specificity protein 1 (Sp1), histone deacetylase 1 (HDAC1), epigenetics, acetylation, ganglioside, gene regulation, B4Galnt1, GM2-synthase, renal cell carcinoma, transcriptional repression
Introduction
Gangliosides are sialic acid–containing glycosphingolipids, ubiquitous in mammalian cells, synthesized by glycosyltransferases. Several gangliosides and corresponding glycosyltransferases are aberrantly expressed in different tumors (1–3), which has been linked with modulation of the tumor microenvironment including but not limited to immune dysfunction (4, 5). Alongside, glycosyltransferases are known to play complex and differential roles in tumor cell behavior (6–8) as well. Recent reports from our laboratory not only linked tumor-derived ganglioside GM2 with increased migration and invasion (9), but also indicated the potential role of GM2 in inducing epithelial-mesenchymal transition (10). Although Human Protein Atlas database demonstrates enhanced mRNA expression of GM2-synthase gene (GalNAcT, B4galnt1) in several cancers including renal cell carcinoma (RCC)2 (5, 11, 12), it remains unclear how GM2-synthase gene transcription is regulated in cancer. This prompted us for a detailed investigation to find out whether epigenetic regulation of GM2-synthase gene occurs in cancer and, if so, how.
There was no direct evidence of epigenetic regulation of GM2-synthase gene until Suzuki et.al. (13) reported histone acetylation to regulate GM2-synthase gene during different stages of mouse brain development. Here, we show for the first time that the transcriptional activity of GM2-synthase in human RCC is regulated by epigenetic mechanisms. Epigenetic marks, such as histone acetylation and DNA methylation, are tissue- and cell-type–specific programs in controlling gene transcription (14, 15). Histone acetylations at H3K9 and H3K14 positions around the TSS are generally associated with active genes (16). Histones are deacetylated by a group of non–DNA-binding proteins, histone deacetylases (HDACs), recruited to gene promoters by several mechanisms, including direct interaction with transcription factors like Sp1 (17–19). Interestingly, studies show that Sp1 can act both as a transcriptional activator or repressor depending on the elements it binds at the promoter of the target gene (20–22).
Toward understanding the molecular mechanism underlying the deregulation of GM2-synthase mRNA expression in RCC, we analyzed its epigenetic regulation in RCC cell lines and primary tumors. We confirmed that de-repression of GM2-synthase gene in RCC cell lines and primary tumors is contributed by higher histone acetylations and lower binding of Sp1-HDAC1 repressor complex at +38/+187 region near TSS (hereafter referred to as Region P).
Results
Increased histone acetylations at Region P of GM2-synthase gene associates with higher GM2-synthase mRNA expression in various cancer cell lines and RCC patient tumor
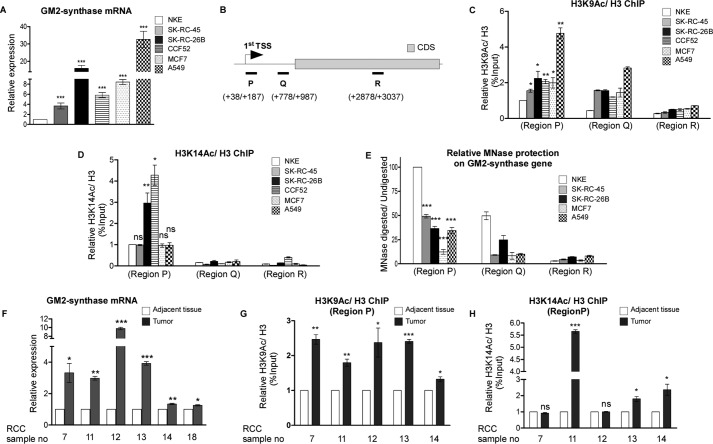
Over-expression of the ganglioside biosynthetic enzyme GM2-synthase mRNA and its corresponding ganglioside GM2 was previously reported in different cancer cell lines as well as patient tumors and also shown in Human Protein Atlas database (4, 5, 9, 23). Expression of GM2-synthase mRNA and corresponding GM2 was assessed in NKE (noncancerous renal epithelial cell), SK-RC-45, SK-RC-26B (renal cell carcinoma), CCF52 (glioblastoma), MCF-7 (breast cancer), and A549 (lung adenocarcinoma) cell lines. Data show elevated but differential levels of GM2-synthase mRNA in the entire cancer cell lines compared with NKE as shown by real-time PCR (Fig. 1A). Expression of ganglioside GM2, as assessed by immunostaining followed by confocal microscopy, reveals elevated levels of GM2 in most cancer cells compared with NKE, except MCF-7 (Fig. S1B). Because the mechanism underlying an aberrant deregulation of GM2-synthase transcription in cancer has not been investigated, we wanted to see whether activation of GM2-synthase mRNA expression in cancer cells/tissues might be associated with increased histone acetylation near the TSS, as only a handful of studies claim that ganglioside synthase genes are epigenetically regulated (13, 24, 25). To address this question, we performed chromatin immunoprecipitation (ChIP) assay with anti–acetyl-H3K9 (H3K9Ac), anti–acetyl-H3K14 (H3K14Ac), and anti-H3 antibodies, followed by quantitative real-time PCR (qPCR).
Figure 1.
Increased histone acetylations at Region P of GM2-synthase gene associate with higher GM2-synthase mRNA expression in various cancer cell lines and RCC patient tumors. A, GM2-synthase mRNA is over-expressed in cancer cell lines. Total RNA was isolated from indicated cell lines and reverse-transcribed. cDNAs were subjected to qPCR using GM2-synthase primer. Relative expression values were normalized to the GAPDH transcript levels and represented as -fold change with respect to NKE. The data represent three independent determinations (average ± S.E.; Student's t test; ***, p < 0.001). B, schematic representation showing three regions along the GM2-synthase gene, namely Region P (+38/+187), Region Q (+778/+987), and Region R (+2878/+3037). C and D, Region P of GM2-synthase gene in cancer cell lines shows higher histone acetylations. ChIP assay performed with different cell lines using antibodies specific for H3, acetyl-H3K9, and acetyl-H3K14. Precipitated chromatin DNA was estimated by qPCR. The results are expressed as relative -fold change with respect to Region P of GM2-synthase gene in NKE. Error bars represent mean ± S.E. of three independent determinations for Region P; Student's t test; *, p < 0.05; **, p < 0.01 versus NKE cells. E, Region P of GM2-synthase gene in cancer cell lines shows lower MNase protection. The MNase protection for each cell line was determined by normalizing the amount of MNase-digested qPCR product to that of the undigested product by using ΔΔCt method (y axis) and represented as relative -folds with respect to Region P of GM2-synthase gene in NKE with an arbitrary value of 100. Error bars represent mean ± S.E. of five independent determinations; Student's t test; ***, p < 0.001 versus Region P of NKE cells. F, GM2-synthase mRNA is over-expressed in RCC tumors. qPCR performed with GM2-synthase primer followed by normalization with β-tubulin and represented as -fold change with respect to corresponding tumor-adjacent tissue. The data represent three independent determinations (average ± S.E.; Student's t test; *, p < 0.05; **, p < 0.01; ***, p < 0.001). G and H, Region P of GM2-synthase gene in tumor tissues shows higher histone acetylations. ChIP assay performed with human RCC tumor and corresponding tumor-adjacent tissues using antibodies specific for H3, acetyl-H3K9, and acetyl-H3K14. Precipitated DNA was PCR amplified as indicated in C and D. Error bars represent mean ± S.E. of three independent determinations; Student's t test; *, p < 0.05; **, p < 0.01; ***, p < 0.001 versus adjacent tissue. ns, not significant.
As histone activation marks like H3K9 acetylation are generally enriched just after the TSS of active genes (26, 27), primers were designed to amplify Region P (+38/+187), Region Q (+778/+987), and Region R (+2878/+3037) of GM2-synthase gene, as represented in Fig. 1B. Indeed, ChIP results demonstrate that relative histone H3K9 (Fig. 1C) as well as H3K14 (Fig. 1D) acetylations are relatively higher at Region P (near to TSS) of GM2-synthase gene compared with the downstream Regions Q and R in all the cell lines, suggesting the specificity of histone acetylations (H3K9 and H3K14) at the Region P of GM2-synthase gene. ChIP assay further confirmed that all the cancer cells have significantly higher histone H3K9 acetylation at Region P compared with NKE (Fig. 1C), which correlates with GM2-synthase mRNA expression profile (Fig. 1A). Increased H3K14 acetylation at Region P was also found in SK-RC-26B and CCF52 relative to NKE (Fig. 1D). Next, micrococcal nuclease (MNase), which cleaves nucleosome-free and linker DNA, was used to evaluate the state of chromatin compaction (28, 29). Results indicate overall highest MNase protection at Region P of GM2-synthase as compared with Regions Q and R, in all the cell lines (Fig. 1E), suggesting that the chromatin at Region P is more compact than Regions Q and R. Results show that Region P of active GM2-synthase genes in the cancer cell lines have significantly lower MNase protection (as evidenced from the ratio of MNase digested to undigested product) relative to the repressed GM2-synthase state in NKE (Fig. 1E), suggesting chromatin compaction at Region P of NKE represses GM2-synthase mRNA expression.
We then investigated the GM2-synthase mRNA expression in clinically characterized RCC tumors and corresponding adjacent tissue. We observed higher GM2-synthase mRNA expression in six RCC tumor tissues over corresponding tumor-adjacent tissues from 22 patients' tissue samples screened (Fig. 1F). Among the six samples, we randomly selected five samples for histone acetylation status at Region P. Indeed our results confirmed H3K9 acetylation was significantly higher in all the tumor tissues over the corresponding adjacent tissues (Fig. 1G), whereas H3K14 acetylation was higher only in three tumor tissues (Fig. 1H). Altogether, these results demonstrate higher histone acetylations (H3K9/H3K14/both) and lower MNase protection at Region P of GM2-synthase gene are associated with higher GM2-synthase mRNA expression in RCC cell lines and primary tumor tissues.
Alteration of histone acetylations at Region P of GM2-synthase gene directly regulates its transcription
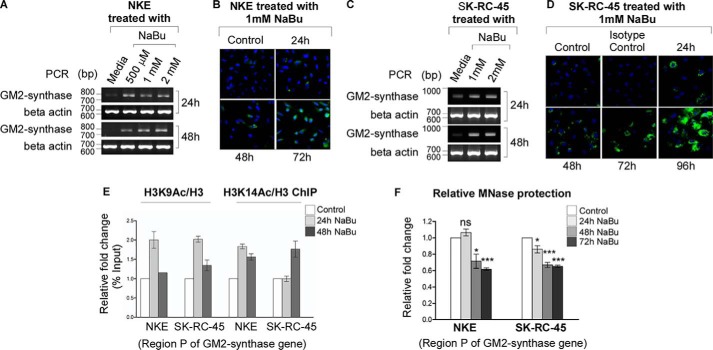
To prove that GM2-synthase mRNA expression is regulated by histone acetylation, we studied the expression of GM2-synthase mRNA and ganglioside GM2 in presence of HDAC inhibitor. We observed increased GM2-synthase mRNA expression following treatment with the HDAC inhibitor sodium butyrate (NaBu) in low GM2-synthase expressing NKE (Fig. 2A) as well as moderate GM2-synthase expressing SK-RC-45 cells (Fig. 2C). Consequently, time-dependent increase in ganglioside GM2 levels were also observed in both cell lines treated with 1 mm NaBu (Fig. 2, B and D). Further, to confirm that NaBu-mediated increase in GM2-synthase transcription involves direct involvement of histone acetylations (H3K9 and H3K14) at Region P, ChIP assay was performed. Results shown in Fig. 2E confirm butyrate-induced GM2-synthase mRNA activation in NKE and SK-RC-45 cell lines involves increased histone H3K9 and H3K14 acetylations. Consequent time-dependent decrease in DNA protection toward MNase digestion was also observed at Region P (Fig. 2F), suggesting chromatin relaxation near the TSS. These results conclusively prove direct involvement of histone acetylations at Region P of GM2-synthase gene with its mRNA expression in RCC cell lines and normal renal NKE cells.
Figure 2.
Alteration of histone acetylations at Region P of GM2-synthase gene directly regulate its transcription. A and C, butyrate increases GM2-synthase mRNA expression. Total RNA was isolated from NKE and SK-RC-45 cell lines upon treatment with indicated concentration of NaBu for different time points and reverse-transcribed. cDNAs were subjected to semi-quantitative PCR using GM2-synthase primer (786 bp). β-actin (687 bp) was used as an internal control for the above PCR experiments (bp). B and D, butyrate increases ganglioside GM2 levels. Immunostaining was performed with hamster anti-human GM2 antibody or hamster IgG antibody (isotype control) as indicated upon time-dependent NaBu treatment on NKE, SK-RC-45. E, ChIP assay performed with different cell lines using antibodies specific for H3, acetyl-H3K9, and acetyl-H3K14. Precipitated chromatin DNA was estimated by qPCR. Error bars represent mean ± S.E. of two independent determinations. F, butyrate decreases chromatin compaction at Region P. Following time-dependent treatment of 1 mm NaBu in NKE and SK-RC-45 cells, loss of chromatin compaction was measured by MNase protection assay. Data from three independent determinations represented as relative -folds with respect to Region P of GM2-synthase gene of untreated control cells (average ± S.E.; Student's t test; *, p < 0.05; ***, p < 0.001). ns, not significant.
DNA methylation is absent at the −73 to +158 region of GM2-synthase gene in normal and RCC cell lines
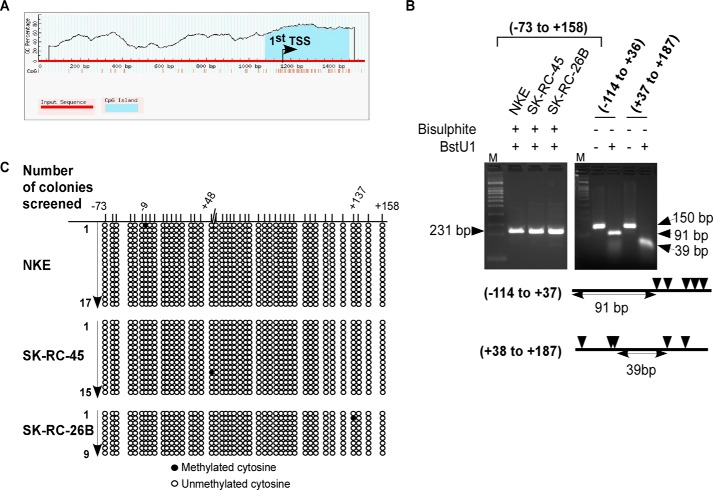
We observed a CpG island around the TSS of GM2-synthase gene encompassing Region P using “MethPrimer” (30) (Fig. 3A). DNA methylations within CpG island of gene promoter leads to gene silencing (31). Therefore, we screened for CpG methylations around the TSS of GM2-synthase gene both in normal NKE and RCC cell lines (SK-RC-45 and SK-RC-26B). BstUI enzyme cleaves 5′-CGCG-3′ DNA sequences. We observed that BstUI enzyme did not cleave the DNA encompassing the genomic region −73/+158 with respect to the TSS of GM2-synthase gene which was pretreated with bisulphite in either NKE or RCC cell lines (Fig. 3B, left gel). This result suggests absence of DNA methylation in this genomic locus in the three cell lines, as bisulphite treatment converts unmethylated cytosine to uracil, thereby rendering BstUI digestion ineffective. In fact, at identical condition, BstUI enzyme completely cleaved the non–bisulphite-treated genomic DNA encompassing the regions around the TSS of GM2-synthase gene, confirming the specificity of the assay (Fig. 3B, right gel). The cleaved DNA at lane 3 and lane 5 matched the expected position of intact DNA, because of the absence of CGCG sequence across contiguous 91 bases (of Region −114/+37) and 39 bases (of Region +38/+187), respectively. Further, we used COBRA assay followed by DNA sequencing for high-resolution screening of CpG methylation around the TSS of GM2-synthase gene in NKE and RCC cell lines. Results clearly demonstrate absence of any prominent CpG methylation in this genomic region of GM2-synthase gene (Fig. 3C). Hence, lack of DNA methylation around the TSS of either transcriptionally repressed GM2-synthase gene in NKE or its active state in RCC cell lines suggests that DNA methylation has no role in epigenetic regulation of renal GM2-synthase gene, at least in this region of our study.
Figure 3.
DNA methylation is absent at the region −73 to +158 region of GM2-synthase gene in normal and RCC cell lines. A, Region P is located within a CpG island. Schematic representation of predicted CpG island using MethPrimer database around the first TSS of GM2-synthase gene encompassing Region P. B, DNA methylation is absent at the −73 to +158 region confirmed by BstUI digestion assay. Isolated genomic DNA from indicated cell lines were treated with bisulphite (left gel) or untreated (right gel). Following PCR, DNA samples were digested with BstUI restriction enzyme. Restriction sites of BstUI within the DNA are indicated by arrowheads (not to scale). The primers used to amplify genomic regions of GM2-synthase gene are listed in Table S1H. C, DNA methylation is absent at the region −73 to +158 region confirmed by bisulphite sequencing. Methylation status of 17 separate TA clones of NKE, 15 and 9 separate TA clones of SK-RC-45 and SK-RC-26B, respectively, were determined by bisulphite sequencing analysis. The filled or open circles represent the methylated or unmethylated CpG sites, respectively, and each horizontal column designates the arbitrary position of a CpG dinucleotide around the TSS of GM2-synthase gene.
The Sp1-binding site within Region P of GM2-synthase gene functions as a repressor
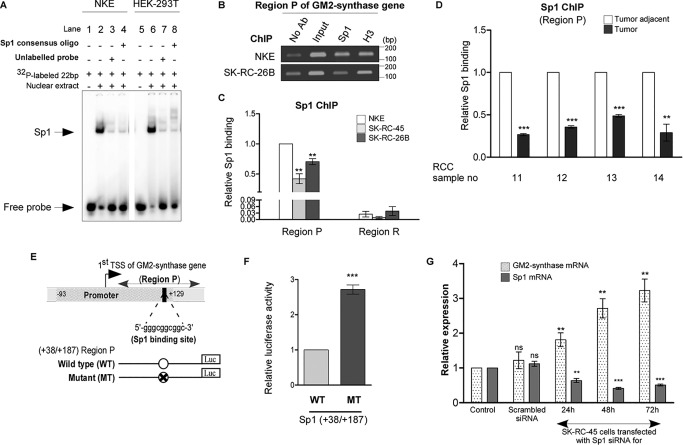
We further investigated the involvement of a possible transcription factor associated with Region P in regulation of GM2-synthase transcription. Previous report predicted one Sp1-binding site within the Region P of GM2-synthase gene (32); however, its role in transcriptional control of GM2-synthase gene is unknown. Here, we identified a +109/+125 Sp1-binding site within Region P using Genomatix's MatInspector (Fig. S2A). To prove that Sp1 binds at +109/+125 region in vitro, we designed a 22-bp oligonucleotide for electrophoretic mobility shift assay (EMSA) which encompasses the Sp1-binding site of GM2-synthase gene. Fig. 4A shows that nuclear lysates of NKE and HEK-293T (lanes 2 and 6) caused a band shift which was competed out not only by using excess (100×) of this unlabeled probe (lanes 3 and 7) but also by a separate unlabeled 22-bp Sp1 consensus oligonucleotide (Santa Cruz Biotechnology), suggesting that Sp1 binding at the +109/+125 is specific in vitro. Next, ChIP assay was performed to assess relative binding of Sp1 at the Region P in low GM2-synthase expressing NKE versus higher GM2-synthase expressing SK-RC-45 and SK-RC-26B. ChIP data revealed that relative binding of Sp1 to Region P (encompassing the +109/+125 Sp1-binding site) was higher than downstream Region R (lacks any Sp1-binding site) in all three cell lines studied, suggesting specific association of Sp1 with Region P (Fig. 4C). Interestingly, we observed that higher binding of Sp1 with Region P in NKE cells relative to the RCC cell lines (SK-RC-26B and SK-RC-45) (Fig. 4, B and C), even though the DNA sequences around the TSS of the gene are identical as confirmed by DNA sequencing (Fig. S1A), suggesting a possible repressor role of Sp1 in GM2-synthase gene transcription. Further, ChIP assay with selected RCC patient samples confirmed higher binding of Sp1 with Region P of repressed GM2-synthase gene in tumor-adjacent tissues relative to corresponding tumors (Fig. 4D). However, expression of Sp1 mRNA in renal cell lines as well as patient samples showed no direct correlation with Sp1 occupancy at Region P of GM2-synthase gene (Fig. S2, B and C). Therefore, to understand the transcriptional activity of the +109/+125 Sp1-binding site within Region P, luciferase reporter assay was performed with WT and Sp1-binding site mutated (MT) Region P of GM2-synthase gene (Fig. 4E) in high GM2-synthase expressing human embryonic kidney (HEK-293T) cells (Fig. S2, D and E). Interestingly, cells transfected with MT construct show significantly higher luciferase activity compared with those transfected with WT construct (Fig. 4F), thereby confirming that the +109/+125 Sp1-binding site at Region P functions as a transcriptional repressor to GM2-synthase gene. Further, knock down of Sp1 in SK-RC-45 cells shows a time-dependent increase in GM2-synthase mRNA expression (Fig. 4G), suggesting Sp1 represses GM2-synthase transcription in SK-RC-45 cells.
Figure 4.
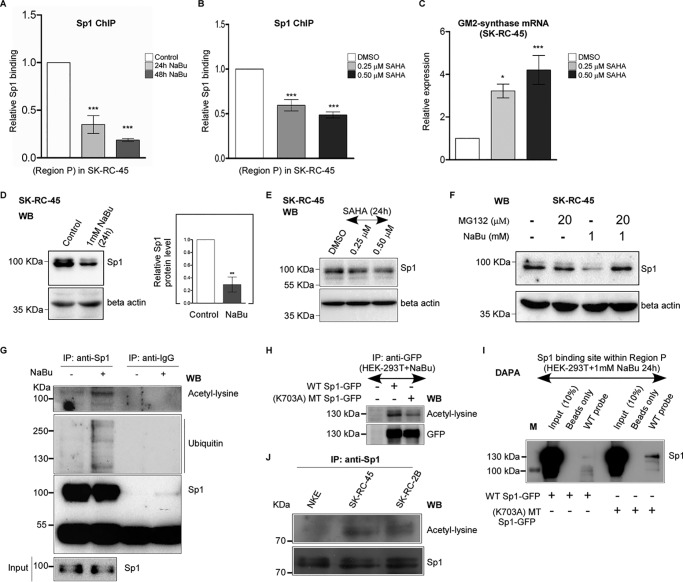
The Sp1-binding site within Region P of GM2-synthase gene promoter functions as a repressor. A, Sp1 binds with Sp1-binding site within Region P in vitro. EMSA used probes as mentioned in Table S1E with nuclear lysates of NKE and HEK-293T cell lines. Interaction of nuclear protein with the [γ-32P]ATP–labeled probes resulted in significant shift (arrow mark) (lanes 2 and 6) which was self-competed with excess of unlabeled probe (lanes 3 and 7) as well as cross-competed with unlabeled 22-bp Sp1 consensus oligonucleotide (lanes 4 and 8). B, relative high Sp1 binding to Region P in transcriptionally repressed state of GM2-synthase gene in NKE compared with transcriptionally active state in RCC cell lines. ChIP assay was done using antibodies specific for Sp1. ChIP assay with antibody specific for H3 and no antibody served as positive and negative controls, respectively. Representative gel image showing immune-precipitated DNA from indicated cell lines was PCR amplified using primers (150 bp) targeting Region P (bp). C, ChIP assay followed by qPCR showing relative Sp1 binding at Region P in the mentioned cell lines. Region R having no Sp1-binding site was used as a negative control. The data represent three independent determinations (average ± S.E.; Student's t test; **, p < 0.01). D, relative high Sp1 binding to Region P with transcriptionally repressed state of GM2-synthase gene in human RCC tumor-adjacent tissues. ChIP assay followed by qPCR for Sp1 binding at Region P was carried out in the human RCC tumor and corresponding tumor-adjacent tissues. The results are expressed as relative -fold change with respect to Region P of corresponding tumor-adjacent tissues. Error bars represent mean ± S.E. of three independent determinations; Student's t test; **, p < 0.01; ***, p < 0.001. E, map of Region P of GM2-synthase gene used for luciferase reporter assay. Schematic representation of the first TSS of GM2-synthase gene showing the core promoter (region −93/+129) as analyzed using Genomatix PromoterInspector, encompassing the +109/+125 Sp1-binding site. The position of Region P is marked. Map of luciferase constructs with Region P are shown along with mutated Sp1-binding site. F, Sp1-binding site at Region P functions as a repressor site. HEK-293T cells were transiently co-transfected with WT-Sp1 or MT-Sp1 firefly luciferase constructs and pRLCMV plasmid. Protein lysates were prepared for Dual-Luciferase Assay. The data represent three independent determinations (average ± S.E., Student's t test, ***, p < 0.001). G, Sp1 knock down in SK-RC-45 cells increases GM2-synthase transcription. SK-RC-45 cells were transfected with siRNA directed against Sp1 mRNA for indicated time points. Scrambled siRNA treatment was done for 72 h. Total RNA was isolated and reverse-transcribed. cDNAs were subjected to qPCR using GM2-synthase primer. Relative expression values were normalized to the housekeeping gene tubulin transcripts levels and represented as -fold change with respect to nontransfected cells as control. The data represent three independent determinations (average ± S.E.; Student's t test; **, p < 0.01; ***, p < 0.001). ns, not significant.
Butyrate induces loss of Sp1 binding with Region P of GM2-synthase gene
To understand the reason behind relative Sp1-binding loss at Region P in high GM2-synthase expressing conditions, we studied the effect of butyrate on Sp1 binding at Region P in SK-RC-45 cells. We investigated the relative binding of Sp1 at Region P in SK-RC-45 cells during transcriptional activation of GM2-synthase gene. Our previous result showed that butyrate increased GM2-synthase mRNA expression in SK-RC-45 cells (Fig. 2C). Hence, we anticipated reduced binding of Sp1 at Region P in SK-RC-45 cells treated with NaBu, as the Sp1-binding site within Region P acts as a transcription repressor (Fig. 4, E and F). Indeed, using ChIP assay we confirmed that NaBu decreased Sp1 binding from Region P in SK-RC-45 cells at time points (Fig. 5A) corresponding to increased GM2-synthase mRNA expression (Fig. 2C), thereby suggesting further that Sp1 may be a part of a transcription repressor complex associated with Region P. To gain an insight into the possible reason behind butyrate-induced loss of Sp1 binding to Region P in SK-RC-45 cells, we studied the effect of butyrate on Sp1 expression. Interestingly, we observed NaBu reduced the protein level of Sp1 (Fig. 5D) but not its mRNA expression (Fig. S3A), suggesting degradation of Sp1 protein levels upon butyrate treatment.
Figure 5.
Butyrate and SAHA induce loss of Sp1 binding with Region P of GM2-synthase gene. A, butyrate treatment reduces Sp1 binding at Region P of GM2-synthase gene. ChIP assay performed with SK-RC-45 cells treated with 1 mm NaBu at mentioned time points using antibody specific for Sp1. Precipitated chromatin DNA was estimated by qPCR. Error bars represent mean ± S.E. of three independent determinations for Region P (average ± S.E.; Student's t test; ***, p < 0.001). B, SAHA treatment reduces Sp1 binding at Region P of GM2-synthase gene. ChIP assay performed with SK-RC-45 cells treated with SAHA at mentioned concentrations for 24 h using antibody specific for Sp1. Precipitated chromatin DNA was estimated by qPCR. Error bars represent mean ± S.E. of three independent determinations for Region P (average ± S.E.; Student's t test; ***, p < 0.001). C, SAHA treatment in SK-RC-45 cells increases GM2-synthase transcription. SK-RC-45 cells were treated with SAHA at mentioned concentrations for 24 h. Total RNA was isolated and reverse-transcribed. cDNAs were subjected to qPCR using GM2-synthase primer. Relative expression values were normalized to the housekeeping gene GAPDH transcripts levels and represented as -fold change with respect to DMSO-treated cells as vehicle control. The data represent three independent determinations (average ± S.E.; Student's t test; *, p < 0.05; ***, p < 0.001). D, NaBu treatment on SK-RC-45 cells down-regulates endogenous Sp1 protein expression. SK-RC-45 cells were treated with 1 mm NaBu for 24 h, and cell extracts were prepared followed by Western blot analysis with antibodies against Sp1 and β-actin. Error bars represent mean ± S.E. of four independent determinations (average ± S.E.; Student's t test; **, p < 0.01). WB, Western blotting. E, SAHA treatment on SK-RC-45 cells down-regulates endogenous Sp1 protein expression. SK-RC-45 cells were treated with SAHA at mentioned concentrations for 24 h, and cell extracts were prepared followed by Western blot analysis with antibodies against Sp1 and β-actin. F, MG132 treatment on SK-RC-45 cells blocks NaBu-mediated down-regulation of Sp1 protein. SK-RC-45 cells were treated with 1 mm NaBu for 24 h, and 20 μm MG132 was added 16 h post NaBu treatment and incubated for remaining 8 h, following which cell extracts were prepared for Western blotting analysis with antibodies against Sp1 and β-actin. G, Sp1 protein in SK-RC-45 cells treated with 1 mm NaBu for 16 h is ubiquitinated and acetylated. Cell extracts were immunoprecipitated (IP) with antibodies specific for Sp1 or normal IgG. Immunocomplexes and input were probed with ubiquitin, pan-acetyl-lysine and Sp1 antibodies. H, mutated Sp1 protein (K703A) shows reduced acetylation compared with WT-Sp1 protein in presence of NaBu. Cell extracts from untransfected HEK-293T or transfected with indicated Sp1 plasmids tagged with GFP were immunoprecipitated (IP) with antibodies specific for GFP. Immunocomplexes were probed with pan-acetyl-lysine and GFP antibodies. I, acetylated WT-Sp1 protein binds less to the Sp1-binding site within Region P of GM2-synthase gene compared with less-acetylated (K703A) mutated Sp1 protein in vitro. DAPA with cell extract of HEK-293T cells which were transfected with either WT-Sp1 or K703A MT-Sp1 plasmids were incubated with 22-bp biotinylated probe encompassing the Sp1-binding site within Region P of GM2-synthase gene (WT probe). Proteins bound to the biotinylated probes were pulled down by streptavidin beads and probed for Sp1 protein using anti-Sp1 antibody by immunoblotting. The molecular mass of over-expressed Sp1 which is tagged with GFP is 125 KDa (Fig. S3B). Beads only panel was included as a negative control. M denotes protein marker. J, Sp1 protein in RCC cell lines shows higher acetylation compared with NKE. Cell extracts were immunoprecipitated (IP) with antibody specific for Sp1. Immunocomplexes were probed with pan-acetyl-lysine and Sp1 antibodies.
Further, to study the role of HDAC inhibitors other than NaBu on GM2-synthase gene activation in SK-RC-45 cells, we chose two potential HDAC inhibitors, namely SAHA and trichostatin A (TSA). Interestingly, treatment of both SAHA as well as TSA led to significant increase in GM2-sythase mRNA level (Fig. 5C and Fig. S4B) as confirmed by qPCR assays, whereas Sp1 protein expression decreased in a dose-dependent manner (Fig. 5E and Fig. S4A), as confirmed by Western blotting. These results suggest that HDAC inhibitors like SAHA and TSA, similar to NaBu (Fig. 5B), induce loss in Sp1 protein expression which potentially may contribute to GM2-synthase gene activation. Moreover, to directly address, whether such HDAC inhibitors indeed induce loss of Sp1 binding with Region P of GM2-synthase gene similar to NaBu (Fig. 5A), we analyzed Sp1 association at Region P by ChIP assay in SAHA-treated SK-RC-45 cells. Results in Fig. 5B, confirm significant dose-dependent loss of Sp1 association with Region P in SAHA-treated SK-RC-45 cells, suggesting that activation of GM2-synthase gene in SK-RC-45 cells by SAHA is associated with loss of Sp1 binding from Region P of GM2-synthase gene.
Proteasomal pathway acts as a significant mode of protein degradation induced by butyrate (33). Here, we explored the possibility that Sp1 protein may be degraded by NaBu treatment involving the proteasomal pathway. In fact, treatment with the proteasomal blocker, MG132 on SK-RC-45 cells pretreated with NaBu, blocked the degradation of Sp1 protein expression as confirmed by Western blotting (Fig. 5F), indicating that the degradation of Sp1 protein by NaBu was mediated through the proteasomal pathway. Again, Sp1 protein degradation through the proteasome pathway involves ubiquitination (34). Therefore, we investigated the effect of NaBu on ubiquitination of Sp1 in SK-RC-45 cells. Immunoprecipitated protein from butyrate-treated SK-RC-45 cells or untreated condition were separated by SDS-PAGE and immunoblotted with anti-ubiquitin antibody. Results clearly indicate increased ubiquitination in response to NaBu treatment which was immunoprecipitated with Sp1 antibody but not with IgG (Fig. 5G). These results confirm that butyrate-induced degradation of Sp1 in SK-RC-45 cells is accompanied by enhanced protein ubiquitination (Fig. 5G), and this is consistent not only with the observed degradation of Sp1 protein (Fig. 5D), but also the inhibition of degradation by the proteasome inhibitor MG132 (Fig. 5F).
Next, acetylation of Sp1 by butyrate was known to reduce its DNA-binding affinity at promoter of specific genes (35). However, the binding affinity of acetylated Sp1 to GM2-synthase gene was unknown. Hence, we investigated the possibility of Sp1 acetylation in SK-RC-45 cells treated with NaBu and consequently its effect on binding with Region P of GM2-synthase gene. We measured expression of acetylated Sp1 protein in cell lines by immunoprecipitating Sp1 and probing with anti–pan-acetyl-lysine antibody. We observed increased Sp1 acetylation in NaBu-treated SK-RC-45 cells (Fig. 5G) as indicated by the appearance of an acetyl-lysine band co-migrating with Sp1, suggesting that acetylation of Sp1 may further contribute to the loss of its binding with Region P in SK-RC-45 cells treated with NaBu. Therefore, to understand whether acetylated Sp1 has indeed lower binding affinity with Region P of GM2-synthase gene, we constructed recombinant Sp1 mutant (K703A) expression plasmid (MT), as the only site known to be acetylated in Sp1 being Lys-703 (36). Indeed, MT-Sp1 protein showed reduced acetylation compared with WT-Sp1 protein that was over-expressed in HEK-293T cells treated with NaBu (Fig. 5H). Hence, we performed DAPA assay with both NaBu-treated and untreated HEK-293T cells that were transfected with either WT-Sp1 or MT-Sp1 to investigate the relative binding of acetylated (WT) Sp1 versus less-acetylated (MT) Sp1 with Region P. Results of DAPA assay with NaBu treatment (Fig. 5I) shows that MT-Sp1 protein binds more to the Region P of GM2-synthase gene compared with WT-Sp1 protein, suggesting that Sp1 acetylation by NaBu contributes to its reduced binding to Region P of GM2-synthase gene. DAPA assay without NaBu treatment (Fig. S4C) shows no significant difference between WT-Sp1 and MT-Sp1 binding in Region P, suggesting that the point mutation does not affect the intrinsic binding capacity of Sp1 apart from acetyl modification. Taken together, the above results show that both acetylation and degradation of Sp1 protein significantly reduce its binding with Region P in SK-RC-45 cells during NaBu-mediated transcriptional activation of GM2-synthase gene.
We further investigated the endogenous Sp1 acetylation levels in NKE, SK-RC-45, and SK-RC-26B cell lines by immunoprecipitating Sp1 and probing with anti–pan-acetyl-lysine antibody (Fig. 5J). Interestingly, Sp1 acetylation levels in RCC cell lines (SK-RC-45 and SK-RC-26B) were higher compared with NKE, suggesting that the relative loss of Sp1 binding with Region P of GM2-synthase gene in RCC cell lines compared with NKE (Fig. 4C) may be contributed by higher endogenous acetylations of Sp1 protein in these cell lines.
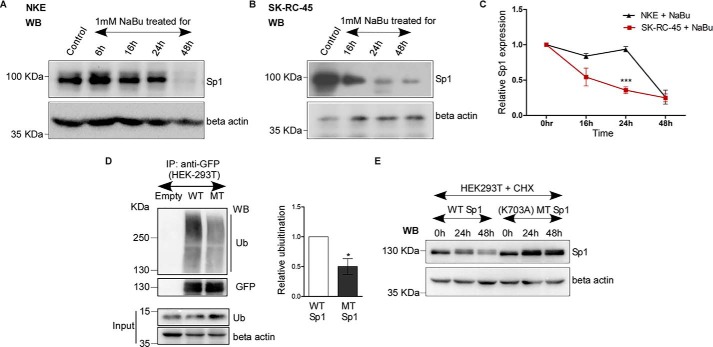
Acetylated Sp1 degrades faster compared with less-acetylated Sp1 in response to butyrate treatment
To understand the relationship between acetylation of Sp1 and its degradation, we investigated the time-dependent loss of Sp1 protein expression by NaBu in NKE and SK-RC-45 cell lines. Fig. 6, A–C shows that the relative loss of Sp1 protein expression in SK-RC-45 cells at 24 h is significantly higher compared with NKE, suggesting that higher Sp1 acetylation in SK-RC-45 cells may contribute to faster degradation by NaBu compared with less-acetylated Sp1 in NKE cells.
Figure 6.
Acetylated Sp1 degrades faster compared with less-acetylated Sp1 in response to butyrate treatment. A and B, NaBu treatment on NKE and SK-RC-45 cells down-regulates endogenous Sp1 protein expression. NKE and SK-RC-45 cells were treated with 1 mm NaBu up to 48 h, and cell extracts were prepared followed by Western blot analysis with antibodies against Sp1 and β-actin. WB, Western blotting. C, relative time-dependent loss of Sp1 protein expression by NaBu in SK-RC-45 is higher compared with NKE. Densitometric analysis of Western blotting was performed using ImageJ software. Error bars represent mean ± S.E. of three independent determinations (average ± S.E.; Student's t test; ***, p < 0.001). D, WT-Sp1 protein (WT) shows higher ubiquitination compared with mutated Sp1 (MT) protein (K703A). Cell extracts with IP buffer (see “Experimental Procedures”) from untransfected HEK-293T or co-transfected with indicated Sp1-GFP and ubiquitin expression plasmids in 3:1 ratio were immunoprecipitated (IP) with antibodies specific for GFP. Immunocomplexes and input were probed with ubiquitin, GFP and β-actin antibodies. Error bars represent mean ± S.E. of three independent determinations (average ± S.E.; Student's t test; *, p < 0.05) by densitometric analysis using ImageJ software. E, WT-Sp1 protein shows lower stability compared with Sp1 MT protein (K703A). HEK-293T were co-transfected with indicated Sp1-GFP and ubiquitin expression plasmids in 3:1 ratio. 24 h post transfection cells were treated with 100 μm cycloheximide (CHX) and chased up to next 48 h. Cell extracts were prepared with IP buffer (see “Experimental Procedures”) followed by Western blot analysis with antibodies against Sp1 and β-actin. The molecular mass of over-expressed Sp1 which is tagged with GFP is 125 KDa. WB, Western blotting.
Moreover, we investigated the possible role of the acetylated Lys-703 residue of Sp1 in regulating Sp1 ubiquitylation and stability of the protein. Previously, we observed that removal of Lys-703 in MT-Sp1 reduces the overall acetylation of Sp1 protein compared with WT-Sp1 (Fig. 5H). Hence, we resolved to understand the affinity of the WT-Sp1 versus MT-Sp1 protein toward ubiquitin. Interestingly, by immunoprecipitation assay in HEK-293T cells co-transfected with ubiquitin expression plasmid along with either WT-Sp1 or MT-Sp1, we confirmed that WT-Sp1 protein is significantly more ubiquitinated relative to MT-Sp1 (Fig. 6D), suggesting that higher acetylation of WT-Sp1 protein (Fig. 5H) may contribute to its higher ubiquitination level. Consequently, through cycloheximide chase assay in HEK-293T cells co-transfected with ubiquitin expression plasmid along with either WT-Sp1 or MT-Sp1, we observed that WT-Sp1 degrades faster than MT-Sp1 (Fig. 6E), suggesting higher ubiquitination of WT-Sp1 decreases its stability compared with less ubiquitinated MT-Sp1. Collectively, these results suggest a possible relationship between acetylation of Sp1 and its consequent degradation.
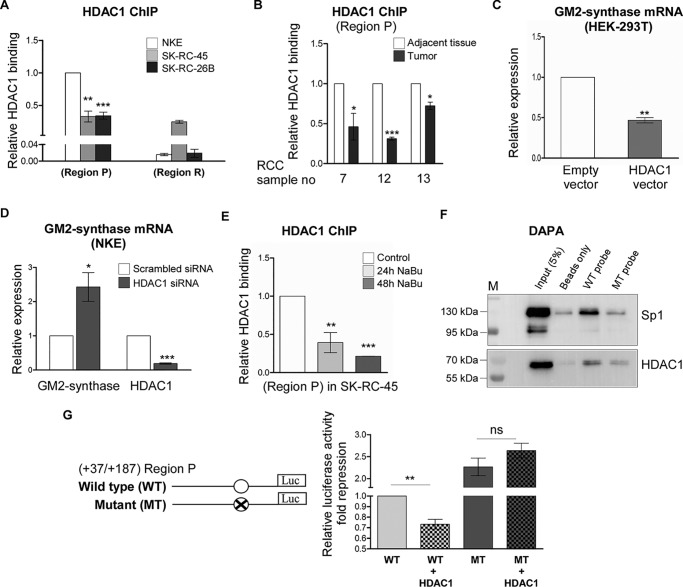
Binding of Sp1 at Region P recruits HDAC1 to repress GM2-synthase transcription
To understand how Sp1 binding at Region P represses GM2-synthase mRNA expression, we checked the possible involvement of the co-repressor HDAC1 in this region. HDAC1 is a non–DNA-binding protein that associates to histones or transcription factors at specific regulatory elements to repress transcription (19). First by ChIP assay, we observed that the relative association of HDAC1 at Region P is higher compared with Region R in all the three cell lines (Fig. 7A), suggesting the binding specificity of HDAC1 to Region P encompassing the Sp1-binding site. Interestingly, at Region P, HDAC1 association was significantly higher in NKE cells relative to RCC cells (Fig. 7A), suggesting that HDAC1 associates more with the transcriptionally repressed GM2-synthase gene. Similar results were obtained in RCC patient tissue samples, where ChIP assay confirmed higher HDAC1 association with Region P of transcriptionally repressed GM2-synthase gene in tumor-adjacent tissues (n = 3) over corresponding tumor tissues (Fig. 7B). Next, to understand whether HDAC1 regulates GM2-synthase transcription, we carried out both over-expression and knock down studies. In fact, transient over-expression of HDAC1 protein in high GM2-synthase expressing HEK-293T cells (Fig. S5, A and B) showed significant reduction of GM2-synthase mRNA level (Fig. 7C), whereas knock down of HDAC1 by siRNA in NKE cells up-regulated GM2-synthase mRNA level (Fig. 7D) as confirmed by qPCR assays, suggesting HDAC1 represses GM2-synthase transcription. Further, to address whether Sp1 recruits non–DNA-binding protein HDAC1 at Region P of GM2-synthase gene, we analyzed HDAC1 association at Region P by ChIP assay in SK-RC-45 cells treated with NaBu. We observed significant loss of HDAC1 association with Region P in NaBu-treated SK-RC-45 cells (Fig. 7E), similar to loss of Sp1 binding (Fig. 5A), suggesting Sp1 and HDAC1 may be part of a transcriptional repressor complex at Region P, whereas NaBu did not decrease HDAC1 protein expression as confirmed by Western blot analysis (Fig. S5C). Moreover, to prove direct involvement of Sp1 in HDAC1 recruitment at Region P, we performed DNA affinity precipitation assay (DAPA). For this, we used biotinylated WT-Sp1 oligo (Sp1 WT) and mutated Sp1 oligo (Sp1 MT) where the +109/+125 Sp1 site is altered, as probes to pull down Sp1 from whole cell lysates of HEK-293T cells, co-transfected with plasmids encoding HDAC1 and Sp1 (Fig. 7F). Results clearly indicate that Sp1 as well as HDAC1 bind more with WT probe over MT probe, thereby indicating that Sp1 binding at Region P is essential for recruitment of HDAC1. Furthermore, luciferase assay performed with lysates of HEK-293T cells co-transfected with WT luciferase constructs encompassing Region P, along with either HDAC1 expression plasmid or empty vector showed significant repression of luciferase activity of WT construct by HDAC1, but interestingly, HDAC1 failed to repress the luciferase activity of the construct having Sp1 site MT (Fig. 7G), demonstrating that Sp1 binding to Region P of GM2-synthase gene recruits HDAC1 to mediate GM2-synthase transcriptional repression.
Figure 7.
Binding of Sp1 at Region P recruits HDAC1 to repress GM2-synthase transcription. A, HDAC1 associates more with transcriptionally repressed GM2-synthase gene in NKE. ChIP assay followed by qPCR for HDAC1 binding at Region P was carried out in the mentioned cell lines. Region R binding was used as negative control. The data represent three independent determinations (average ± S.E.; Student's t test; **, p < 0.01; ***, p < 0.001 versus NKE). B, HDAC1 associates more with transcriptionally repressed GM2-synthase gene in human RCC tumor-adjacent tissues. ChIP assay followed by qPCR for HDAC1 binding at Region P was carried out in the human RCC tumor and corresponding tumor-adjacent tissues. The results are expressed as relative -fold change with respect to Region P of corresponding tumor-adjacent tissues. Error bars represent mean ± S.E. of three independent determinations; Student's t test; *, p < 0.05; ***, p < 0.001. C, GM2-synthase mRNA expression is down-regulated in HEK-293T upon transient over-expression of HDAC1. HEK-293T cells were transiently transfected with HDAC1 or empty plasmids as control. 60 h post transfection, total RNA was isolated for reverse transcription. cDNAs were subjected to qPCR using GM2-synthase primer. Relative expression values were normalized to the GAPDH transcripts levels and represented as -fold change with respect to control. The data represent three independent determinations (average ± S.E.; Student's t test; **, p < 0.01). D, HDAC1 knock down increases GM2-synthase transcription in NKE cells. NKE cells were transfected with siRNA directed against HDAC1 mRNA or scrambled siRNA oligo as control. GM2-synthase was qPCR amplified as mentioned in C. The data represent four independent determinations (average ± S.E.; Student's t test; *, p < 0.05; ***, p < 0.001). E, butyrate treatment reduces HDAC1 association at Region P of GM2-synthase gene. ChIP assay performed with SK-RC-45 treated with 1 mm NaBu at mentioned time points using antibody specific for HDAC1. Precipitated chromatin DNA was estimated by qPCR. Error bars represent mean ± S.E. of three independent determinations for Region P (average ± S.E.; Student's t test; **, p < 0.01; ***, p < 0.001). F, Sp1 recruits HDAC1 to the Sp1-binding site of Region P. Nuclear extract of HEK-293T cells transfected with WT-Sp1 and HDAC1 plasmids was analyzed by DAPA, using 22-bp biotinylated probes designed from the Region P of GM2-synthase gene encompassing Sp1-binding site (Sp1 WT probe) and corresponding Sp1 site mutated oligo (Sp1 MT probe). Proteins binding to the biotinylated probes were pulled down by streptavidin beads and probed for Sp1 and HDAC1 proteins with indicated antibodies by immunoblotting. Beads only panel was included as a negative control. G, HDAC1 fails to repress luciferase activity of Region P construct encompassing mutated Sp1-binding site. Map of luciferase constructs showing WT- and MT-Sp1–binding site encompassing the Region P of GM2-synthase gene. HEK-293T cells were transiently co-transfected with WT-Sp1 or MT-Sp1 luciferase constructs along with HDAC1 or empty plasmids as control. Protein lysates were prepared for luciferase assay. The data were expressed as -fold induction relative to HEK-293T cells co-transfected with empty vector and WT-Sp1 construct and represented from five independent determinations (average ± S.E.; Student's t test; ns, not significant; **, p < 0.001).
Discussion
GM2-synthase is a key enzyme in the ganglioside biosynthesis pathway (37). The aberrant over-expression of GM2-synthase mRNA expression in various cancers is well-established (Human Protein Atlas). Increased GM2-synthase mRNA expression and consequent over-expression of ganglioside GM2 have been found to increase cancer cell migration as well as suppression of specific host immune response (9, 23). Recently, analysis from The Cancer Genome Atlas dataset reveals RCC patients with high GM2-synthase mRNA expression shows poorer prognosis (Human Protein Atlas). However, the GM2-synthase gene regulation in RCC is poorly understood. Here, for the first time we report an epigenetic regulation of GM2-synthase gene in RCC. We showed that activated state of GM2-synthase gene in cancer cells is regulated by higher histone acetylations along with decreased Sp1-HDAC1 repressor complex.
Initially, we observed increased transcription of GM2-synthase in various cancer cell lines and primary RCC tumor tissues. It is known that GM2-synthase mRNA expression increases in neuronal cells during development (38). Indeed, in a previous report, epigenetic activation of GM2-synthase gene involving higher histone acetylations was implicated during different stages in mouse brain development (13). We used several strategies to investigate whether histone acetylations and DNA methylation have any role on regulation of GM2-synthase gene in RCC. First, we showed higher histone acetylations (H3K9/H3K14/both) associate with Region P of active GM2-synthase gene in cancer cell lines and primary RCC tumors over normal cell line and tumor-adjacent tissues by ChIP assay. Second, through COBRA assay we showed that the region around the TSS of GM2-synthase gene lacks DNA methylation in both normal NKE and RCC cell lines. Absence of DNA methylation around the TSS in low GM2-synthase expressing cell line NKE indicates the possibility that GM2-synthase gene may not be permanently inactive but rather it is poised for epigenetic activation. Indeed, treatment of HDAC inhibitor (NaBu) activated transcription of GM2-synthase gene in NKE. Third, using HDAC inhibitor (NaBu) on both NKE and SK-RC-45 cells, we established that increased histone acetylations at Region P transcriptionally activates GM2-synthase gene.
Previously, a report on human GM2-synthase promoter predicted an Sp1 transcription factor–binding site just after the TSS within Region P. However, its role in GM2-synthase regulation was unknown. Our ChIP results revealed that Sp1 protein binds more with repressed GM2-synthase gene in cell lines and tissue samples. Further, siRNA-mediated knock down of Sp1 increased GM2-synthase transcription in SK-RC-45 cells. Consistent with these data, luciferase data showed increased luciferase activity in cells transfected with mutated Sp1-binding site, confirming the repressor function of this site. Moreover, NaBu-mediated transcriptional activation of GM2-synthase gene in SK-RC-45 cells showed significant loss of Sp1 binding from Region P, further suggesting association of Sp1 at Region P represses transcription of GM2-synthase gene. We report here that butyrate-induced loss of Sp1 binding from Region P in SK-RC-45 cells is contributed by proteasome-dependent degradation and acetylation of Sp1 protein. Furthermore, using another potential HDAC inhibitor, SAHA, we observed similar loss of Sp1 protein expression and binding with Region P of GM2-synthase gene in SK-RC-45 cells along with consequent activation of GM2-synthase transcription, suggesting specific HDAC inhibitors induce the loss of Sp1 binding at Region P which contributes to GM2-synthase gene activation in SK-RC-45 cells.
To investigate the possibility that a co-repressor is associated with transcriptional repression of GM2-synthase in normal state, we studied binding of HDAC1 at the Region P of GM2-synthase gene. The HDAC1 protein acts as transcriptional co-repressor and regulates transcription of a number of genes through a variety of ways. First, they act as a bridge connecting different transcription factors and co-repressors to the transcription apparatus (19, 39). Second, histone deacetylase activity of HDAC1 represses transcription by deacetylating nucleosomal histones (40). From our ChIP data showing association of both Sp1 and HDAC1 at Region P of repressed GM2-synthase gene, we propose Sp1 binding at the Region P recruits HDAC1 as part of a repressor complex. To validate such a notion, we examined the Sp1 and HDAC1 binding at Region P of GM2-synthase gene in SK-RC-45 cells upon NaBu-induced GM2-synthase transcriptional activation. Interestingly, NaBu treatment decreased the association of HDAC1 without any change in its protein expression, suggesting its association with Region P depends on Sp1 binding. Also, activation of GM2-synthase transcription was observed under HDAC1 knock down condition whereas the reverse was observed under HDAC1 over-expressed condition. Further, results of DAPA and luciferase assay with Sp1-binding site mutated constructs revealed that Sp1 binding at the Region P is a prerequisite for recruitment of HDAC1 in this region. Collectively, these data indicate that association of HDAC1 at Region P down-regulates GM2-synthase mRNA expression.
The results presented in this study may have important implication in regulating GM2-synthase transcription in cancer cells by targeting Sp1-HDAC1 complex specifically at Region P. This finding also provides the gateway for translating this in animal models, using either syngeneic mouse tumor model or xenografts with engineered tumor cells, where deacetylated Sp1-HDAC1 is targeted to the GM2-synthase TSS to achieve targeted transcriptional repression in cancer cells leading to a favorable response. However, this is a subject of an independent study beyond the scope of the present one, but holds tremendous potential as a possible targeted approach in blocking GM2-synthase mRNA expression in animal models.
Experimental procedures
Cell culture and transfection
NKE, SK-RC-45, and SK-RC-26B were kind gifts from Dr. James H. Finke (Cleveland Clinic). CCF52 was kindly provided by Dr. Michael Vogelbaum (Cleveland Clinic). A549, HEK-293T, and MCF7 cells were procured from the National Centre for Cell Science, Pune, India. All the above cell lines were cultured in complete RPMI 1640 except A549 and HEK-293T which were cultured in DMEM using standard procedures (9). Cells were transfected with indicated plasmids using Lipofectamine-LTX (Life Technologies). RNAi was achieved by using 100 nm siRNA (pool of four siRNA duplexes) of Sp1 and HDAC1 (SMARTpool reagent, Dharmacon, Lafayette, CO). A scrambled siRNA (nontargeting siRNA, Dharmacon) was used as a control for potential nonspecific effects caused by siRNA transfection. Cell lines were transfected with Lipofectamine 2000 (Life Technologies) and harvested at indicated time points after transfection. The following drugs/inhibitors were used: Sodium butyrate (Sisco Research Laboratories, India), MG132, SAHA, and trichostatin A (Sigma-Aldrich), cycloheximide (Sisco Research Laboratories, India).
Human RCC tissue samples
Postsurgical RCC tumor tissue samples and corresponding normal adjacent tissue samples (n = 22) were collected from Indian patients. Studies were approved by the institutional human ethical committee of Bose Institute (BIHEC/2017–18/4) and Nil Ratan Sircar Medical College and Hospital, Kolkata (No/NMC/3650) and conducted in compliance with the Declaration of Helsinki principles.
Plasmids and site-directed mutagenesis
pHDAC1-FLAG was a kind gift from Dr. Eric Verdin (Addgene plasmid no. 13820). pHA-ubiquitin was a kind gift from Dr. Edward Yeh (Addgene plasmid no. 18712). pSp1 was a kind gift from Dr. Beatrice Yue (Addgene plasmid no. 39325) and modified prior to use. The primers used to make recombinant Sp1 mutant (K703A) (41) are given in Table S1. Region P (+38/+187) of GM2-synthase gene was amplified from human genomic DNA with the primers listed in Table S1. The amplified regions were cloned into the linearized pTZ57R/T (Fermentas, Lithuania) by the T/A cloning method. The fragments were then subcloned into luciferase reporter vector pGL3 basic (Promega, Madison, WI) using restriction enzymes KpnI and HindIII (New England Biolabs, Beverly, MA). Deletion of Sp1-binding site within Region P was performed by site-directed mutagenesis, using primer sequences given in Table S1. All constructs were verified by sequencing.
Luciferase assay
After transfection, cells were washed with phosphate-buffered saline (PBS) and subsequently lysed with luciferase lysis buffer supplied with the Dual-Luciferase Assay kit (Promega, Madison, WI). After a short vortex, whole cell lysates were centrifuged at 4 °C at 6000 rpm for 5 min, and total protein concentration in each lysate was measured by BCA Protein Assay (Thermo Fisher Scientific). 20 μl of supernatants with equal protein was mixed with 30 μl of Luciferase Assay substrate followed by 30 μl Stop & Glo Buffer. Luminescence was measured in Varioskan Flash multimode reader (Thermo Fisher Scientific). Each assay was performed in triplicate. -Fold activation values were calculated as mean of three separate experiments.
RNA isolation, RT-PCR, and real-time PCR
Total RNA was extracted using TRIzol Reagent (Invitrogen). cDNA was prepared from 1 μg of total RNA using Verso cDNA synthesis Kit (Thermo Scientific). Total cDNA was diluted 1:4 times and 4 μl of cDNA was used for either semi-quantitative PCR or real-time PCR (qPCR). Real-time PCR was performed on the 7500 Fast Real-time PCR system (Applied Biosystems) using power SYBR Green PCR Master Mix (Applied Biosystems). All primers were custom synthesized by Integrated DNA Technologies (IDT), Coralville, IA. The primers used for selected genes were given in Table S1. The comparative threshold cycle method (ΔΔCt) was used to quantify relative amounts of product transcripts with specific housekeeping genes as mentioned in figure legends as endogenous reference control.
DNA affinity precipitation assay (DAPA)
DNA affinity precipitation assay was performed as described earlier (42). For construction of the annealing probes, both sense (biotinylated) and antisense oligonucleotides were synthesized (IDT, Coralville, IA) and annealed following standard protocol. The sequences of the annealing probes used are given in Table S1. Briefly, nuclear extracts from HEK-293T cells transfected with 12 μg of indicated plasmids were precleared with 10 μl of streptavidin-agarose beads (Sigma-Aldrich), and the precleared nuclear extract was used for binding with biotinylated annealing probes as described earlier. Following incubation with 30 μl of streptavidin-agarose beads with rotation for 1 h at room temperature, the beads were washed three times with 0.1% Nonidet P-40 in 1× PBS and eluted with 30 μl of 2× loading dye and then separated by SDS-PAGE followed by Western blot analysis.
Immunoprecipitation and Western blot analysis
For immunoprecipitation assay, IP buffer (50 mm Tris-HCl, pH 7.9, 10 mm KCl, 1 mm EDTA, 0.2% Nonidet P-40, 10% glycerol) and nuclear lysis buffer (20 mm Tris-HCl, pH 7.9, 10 mm KCl, 1 mm EDTA, 20% glycerol, 400 mm NaCl) containing protease and phosphatase inhibitor (Thermo Fisher Scientific) were used or RIPA was used to prepare whole cell lysate for Western blotting. The supernatants were incubated overnight with antibodies specific for Sp1 (D4C3, 9389) (Cell Signaling Technology). Normal rabbit IgG (3900S) (Cell Signaling Technology) was taken as an isotype control for immunoprecipitation. The antibody–protein complex was precipitated with Pierce Protein A Plus Agarose beads (Thermo Scientific) and washed, and subsequently the protein complex was eluted by SDS-lysis buffer. The eluted samples were then processed for Western blot analysis with antibodies specific for ubiquitin (3936) (Cell Signaling Technology) and pan-acetyl-lysine (623401) (BioLegend). Western blotting was performed as described previously (9). The primary antibodies used as mentioned in the figures are HDAC1 (10E2, 5356) (Cell Signaling Technology), FLAG (Sigma-Aldrich), tubulin (BB-AB0119), and GFP (BB-AB0065) (BioBharati, India), β-actin (Imgenex, India). Bands were detected using Clarity Western ECL substrate (Bio-Rad) after treating with HRP-conjugated rabbit (7074P2) or mouse (7076S) secondary antibodies (Cell Signaling Technology).
Immunofluorescent staining
Immunofluorescent staining of the cell lines was done with hamster monoclonal anti-human GM2 antibody (DMF10.167.4, a gift from Dr. Kenneth Rock, University of Massachusetts Medical School, Worcester, MA and Dr. James H. Finke, Cleveland Clinic, Cleveland, OH) by methods described previously (9, 10). In brief, cells were grown on coverslips, fixed with 3.7% paraformaldehyde, washed with 1× PBS and stained with hamster anti-GM2 antibody (1:50) overnight at 4 °C in a humid chamber. Cells were washed with PBS, counterstained with Alexa Fluor-488 goat anti-hamster IgG antibody (1:500) for 1 h at room temperature. Cells were washed with PBS, mounted on slides, and observed using fluorescence microscope (Leica Microsystems India Pvt. Ltd.).
Electrophoretic mobility shift assay
EMSA was done using 22-bp fragment encompassing the Sp1-binding site of GM2-synthase gene and 22-bp Sp1 consensus oligo (Santa Cruz Technology). Approximately, 10 fmol DNA was mixed with 25 μg of nuclear lysate in 1× EMSA buffer (10 mm Tris-HCl, pH 7.5, 5 mm NaCl, 0.1 mm EDTA, 0.5 mm MgCl2, 0.1 mm DTT, 5% glycerol, and 1.25 μg/ml poly(dI-dC)) and the mixture was incubated on ice for 30 min. The reaction mixture was resolved by 5% native PAGE in 0.5× Tris borate-EDTA at 4 °C and analyzed by phosphoimaging. The sequences of the annealing probes used are given in Table S1.
Chromatin immunoprecipitation
Cells were cross-linked with 1% formaldehyde at room temperature for 10 min by gentle shaking followed by treatment with 1 mm glycine for 5 min. Cells were washed twice with ice-cold 1× PBS and scrapped with 1 ml Nonidet P-40 lysis buffer (50 mm Tris HCl, pH 8, 10 mm EDTA, 1% SDS, protease inhibitor) and incubated at 4 °C for 30 min with occasional vortexing. For tissue, frozen samples were thawed on ice and minced. Tissue pieces (100 mg) were cross-linked with 1% formaldehyde as stated above. At this point, cross-linked tissues were re-suspended in Nonidet P-40 lysis buffer and homogenized several times with a 2 ml Dounce homogenizer. Nuclei were collected by centrifuging at 4 °C for 5 min at 720 g. The nuclear pellets were washed twice and re-suspended in 800 μl of MNase digestion buffer (10 mm Tris HCl, pH 7.5, 15 mm NaCl, 60 mm KCl, 0.15 mm spermidine) and incubated at 37 °C for 10 min with MNase (30 units) to achieve average chromatin fragment size of 200 bp. Digestion was stopped by addition of 200 μl 5× nuclear lysis buffer (50 mm Tris HCl, pH 8, 10 mm EDTA, 1% SDS) and centrifuged at 10,000 rpm for 10 min. The supernatant was diluted 10 times with ChIP dilution buffer (1.1% Triton X-100, 1.2 mm EDTA, 16.7 mm Tris-HCl, pH 8, 167 mm NaCl) and precleared. Immunoprecipitation was carried out overnight with antibodies specific for H3 (ab1791), acetyl-histone H3K9 (ab10812), acetyl-histone H3K14 (ab52946) (Abcam), Sp1 (D4C3, 9389), HDAC1 (10E2, 5356) (Cell Signaling Technology). Next day, preblocked Pierce Protein A Plus Agarose beads (Thermo Scientific) were added for binding to pulled chromatin complex. Beads were washed with high-salt buffer, low-salt buffer, LiCl buffer, and TE consecutively for 10 min each. Following RNase A and Proteinase K treatment, the beads were kept overnight for de–cross-linking at 65 °C. Phenol-chloroform extraction followed by ethanol precipitation was performed. The DNA pellet was dissolved in H2O and used for qPCR analysis using gene-specific primers (Table S1).
Micrococcal nuclease (MNase) protection assay
MNase protection assay was performed as described elsewhere (29), with some modifications. The same procedure for ChIP was used until MNase digestion. At this point MNase digestion was terminated using 100 mm EDTA and 10 mm EGTA. The nuclei were processed like ChIP samples from the point of elution from the beads, before resolving the isolated DNA on a 2% agarose gel. The DNA band corresponding to mononucleosome was excised, gel-extracted using a kit (Qiagen) and 20 ng of DNA from MNase-digested and undigested samples was amplified by real time PCR.
Combined bisulphite restriction analysis (COBRA) assay
DNA methylation levels in cell lines were measured by COBRA assay (43) using EZ DNA Methylation-Gold Kit (Zymo Research). The sequences of the primer sets for COBRA assay are given in Table S1. PCR products were cloned in a pTZ57R/T vector, sequence verified using T7 forward primer (Table S1). For BstUI digestion, PCR products were purified with PCR purification kit (Qiagen), digested with BstUI, and detected on 3% agarose gel.
Statistical analysis
Student's t test (paired) was used to determine the p values using GraphPad Prism software. In every case, the value of (*) p < 0.05, (**) p < 0.01, and (***) p < 0.001 were considered to statistically significant.
Author contributions
A. B. data curation; A. B. formal analysis; A. B. validation; A. B., B. M., and K. B. visualization; A. B., B. M., A. D., and K. B. methodology; A. B. and K. B. writing-original draft; A. B., B. M., and K. B. writing-review and editing; T. K. M. resources; K. B. conceptualization; K. B. supervision; K. B. funding acquisition.
Supplementary Material
Acknowledgments
We thank Dr. James H. Finke for kindly providing NKE, SK-RC-45, and SK-RC-26B and Dr. Michael Vogelbaum for CCF52 cell lines. We also thank Dr. Shubho Chaudhuri and Prof. Mahadeb Pal for valuable technical input.
This work was supported by the Bose Institute, Kolkata, India, and the Department of Biotechnology, India Grants BT/PR5338/MED/30/989/2013 and BT/469/NE/TBP/2013. This work was also supported by Senior Research Fellowships from the University Grants Commission (to A. B.) and the Council of Scientific and Industrial Research, India (to B. M.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S5 and Table S1.
- RCC
- renal cell carcinoma
- TSS
- transcription start site
- HDAC
- histone deacetylase
- NKE
- normal kidney epithelial
- qPCR
- quantitative real-time PCR
- MNase
- micrococcal nuclease
- NaBu
- sodium butyrate
- MT
- mutant
- TSA
- trichostatin A
- DAPA
- DNA affinity precipitation assay.
References
- 1. Tsuchida T., Saxton R. E., and Irie R. F. (1987) Gangliosides of human melanoma: GM2 and tumorigenicity. J. Natl. Cancer Inst. 78, 55–60 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 2. Yuyama Y., Dohi T., Morita H., Furukawa K., and Oshima M. (1995) Enhanced expression of GM2/GD2 synthase mRNA in human gastrointestinal cancer. Cancer 75, 1273–1280 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 3. Petretti T., Kemmner W., Schulze B., and Schlag P. M. (2000) Altered mRNA expression of glycosyltransferases in human colorectal carcinomas and liver metastases. Gut 46, 359–366 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chahlavi A., Rayman P., Richmond A. L., Biswas K., Zhang R., Vogelbaum M., Tannenbaum C., Barnett G., and Finke J. H. (2005) Glioblastomas induce T-lymphocyte death by two distinct pathways involving gangliosides and CD70. Cancer Res. 65, 5428–5438 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 5. Biswas S., Biswas K., Richmond A., Ko J., Ghosh S., Simmons M., Rayman P., Rini B., Gill I., Tannenbaum C. S., and Finke J. H. (2009) Elevated levels of select gangliosides in T cells from renal cell carcinoma patients is associated with T cell dysfunction. J. Immunol. 183, 5050–5058 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Manfredi M. G., Lim S., Claffey K. P., and Seyfried T. N. (1999) Gangliosides influence angiogenesis in an experimental mouse brain tumor. Cancer Res. 59, 5392–5397 [PubMed] [Google Scholar]
- 7. Prinetti A., Aureli M., Illuzzi G., Prioni S., Nocco V., Scandroglio F., Gagliano N., Tredici G., Rodriguez-Menendez V., Chigorno V., and Sonnino S. (2010) GM3 synthase overexpression results in reduced cell motility and in caveolin-1 upregulation in human ovarian carcinoma cells. Glycobiology 20, 62–77 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 8. Aoki H., Satoh M., Mitsuzuka K., Ito A., Saito S., Funato T., Endoh M., Takahashi T., and Arai Y. (2004) Inhibition of motility and invasiveness of renal cell carcinoma induced by short interfering RNA transfection of β 1,4GalNAc transferase. FEBS Lett. 567, 203–208 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 9. Kundu M., Mahata B., Banerjee A., Chakraborty S., Debnath S., Ray S. S., Ghosh Z., and Biswas K. (2016) Ganglioside GM2 mediates migration of tumor cells by interacting with integrin and modulating the downstream signaling pathway. Biochim. Biophys. Acta 1863, 1472–1489 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 10. Mahata B., Banerjee A., Kundu M., Bandyopadhyay U., and Biswas K. (2015) TALEN mediated targeted editing of GM2/GD2-synthase gene modulates anchorage independent growth by reducing anoikis resistance in mouse tumor cells. Sci. Rep. 5, 9048 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hoon D. S., Okun E., Neuwirth H., Morton D. L., and Irie R. F. (1993) Aberrant expression of gangliosides in human renal cell carcinomas. J. Urol. 150, 2013–2018 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 12. Hoon D. S., Irie R. F., and Cochran A. J. (1988) Gangliosides from human melanoma immunomodulate response of T cells to interleukin-2. Cell. Immunol. 111, 410–419 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 13. Suzuki Y., Yanagisawa M., Ariga T., and Yu R. K. (2011) Histone acetylation-mediated glycosyltransferase gene regulation in mouse brain during development. J. Neurochem. 116, 874–880 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bonasio R., Tu S., and Reinberg D. (2010) Molecular signals of epigenetic states. Science 330, 612–616 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bannister A. J., and Kouzarides T. (2011) Regulation of chromatin by histone modifications. Cell Res. 21, 381–395 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Karmodiya K., Krebs A. R., Oulad-Abdelghani M., Kimura H., and Tora L. (2012) H3K9 and H3K14 acetylation co-occur at many gene regulatory elements, while H3K14ac marks a subset of inactive inducible promoters in mouse embryonic stem cells. BMC Genomics 13, 424 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Won J., Yim J., and Kim T. K. (2002) Sp1 and Sp3 recruit histone deacetylase to repress transcription of human telomerase reverse transcriptase (hTERT) promoterin normal human somatic cells. J. Biol. Chem. 277, 38230–38238 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 18. Jiang G., Espeseth A., Hazuda D. J., and Margolis D. M. (2007) c-Myc and Sp1 contribute to proviral latency by recruiting histone deacetylase 1 to the human immunodeficiency virus type 1 promoter. J. Virol. 81, 10914–10923 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Doetzlhofer A., Rotheneder H., Lagger G., Koranda M., Kurtev V., Brosch G., Wintersberger E., and Seiser C. (1999) Histone deacetylase 1 can repress transcription by binding to Sp1. Mol. Cell. Biol. 19, 5504–5511 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Li R., Hodny Z., Luciakova K., Barath P., and Nelson B. D. (1996) Sp1 activates and inhibits transcription from separate elements in the proximal promoter of the human adenine nucleotide translocase 2 (ANT2) gene. J. Biol. Chem. 271, 18925–18930 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 21. Zaid A., Hodny Z., Li R., and Nelson B. D. (2001) Sp1 acts as a repressor of the human adenine nucleotide translocase-2 (ANT2) promoter. Eur. J. Biochem. 268, 5497–5503 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 22. Shou Y. P., Baron S., and Poncz M. (1998) An Sp1-binding silencer element is a critical negative regulator of the megakaryocyte-specific αIIb gene. J. Biol. Chem. 273, 5716–5726 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 23. Biswas K., Richmond A., Rayman P., Biswas S., Thornton M., Sa G., Das T., Zhang R., Chahlavi A., Tannenbaum C. S., Novick A., Bukowski R., and Finke J. H. (2006) GM2 expression in renal cell carcinoma: Potential role in tumor-induced T-cell dysfunction. Cancer Res. 66, 6816–6825 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 24. Taki T., Hirabayashi Y., Kondo R., Matsumoto M., and Kojima K. (1979) Effect of butyrate on glycolipid metabolism of two cell types of rat ascites hepatomas with different ganglioside biosynthesis. J. Biochem. 86, 1395–1402 [DOI] [PubMed] [Google Scholar]
- 25. Tsai Y. T., and Yu R. K. (2014) Epigenetic activation of mouse ganglioside synthase genes: Implications for neurogenesis. J. Neurochem. 128, 101–110 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 26. Wang Z., Zang C., Rosenfeld J. A., Schones D. E., Barski A., Cuddapah S., Cui K., Roh T.-Y., Peng W., Zhang M. Q., and Zhao K. (2008) Combinatorial patterns of histone acetylations and methylations in the human genome. Nat. Genet. 40, 897–903 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Krishnakumar R., and Kraus W. L. (2010) The PARP side of the nucleus: Molecular actions, physiological outcomes, and clinical targets. Mol. Cell 39, 8–24 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Krishnakumar R., and Kraus W. L. (2010) PARP-1 regulates chromatin structure and transcription through a KDM5B-dependent pathway. Mol. Cell 39, 736–749 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Petesch S. J., and Lis J. T. (2008) Rapid, transcription-independent loss of nucleosomes over a large chromatin domain at Hsp70 loci. Cell 134, 74–84 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Li L. C., and Dahiya R. (2002) MethPrimer: Designing primers for methylation PCRs. Bioinformatics 18, 1427–1431 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 31. Herman J. G., Latif F., Weng Y., Lerman M. I., Zbar B., Liu S., Samid D., Duan D. S., Gnarra J. R., and Linehan W. M. (1994) Silencing of the VHL tumor-suppressor gene by DNA methylation in renal carcinoma. Proc. Natl. Acad. Sci. U.S.A. 91, 9700–9704 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Furukawa K., Soejima H., Niikawa N., Shiku H., and Furukawa K. (1996) Genomic organization and chromosomal assignment of the human β1,4-N-acetylgalactosaminyltransferase gene. J. Biol. Chem. 271, 20836–20844 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 33. Li C. J., and Elsasser T. H. (2005) Butyrate-induced apoptosis and cell cycle arrest in bovine kidney epithelial cells: Involvement of caspase and proteasome pathways. J. Anim. Sci. 83, 89–97 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 34. Abdelrahim M., and Safe S. (2005) Cyclooxygenase-2 inhibitors decrease vascular endothelial growth factor expression in colon cancer cells by enhanced degradation of Sp1 and Sp4 proteins. Mol. Pharmacol. 68, 317–329 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 35. Waby J. S., Chirakkal H., Yu C., Griffiths G. J., Benson R. S. P., Bingle C. D., and Corfe B. M. (2010) Sp1 acetylation is associated with loss of DNA binding at promoters associated with cell cycle arrest and cell death in a colon cell line. Mol. Cancer 9, 275 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hung J.-J., Wang Y.-T., and Chang W.-C. (2006) Sp1 deacetylation induced by phorbol ester recruits p300 to activate 12(S)-lipoxygenase gene transcription. Mol. Cell. Biol. 26, 1770–1785 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Harlalka G. V., Lehman A., Chioza B., Baple E. L., Maroofian R., Cross H., Sreekantan-Nair A., Priestman D. A., Al-Turki S., and McEntagart M. E. (2013) Mutations in B4GALNT1 (GM2 synthase) underlie a new disorder of ganglioside biosynthesis. Brain 136, 3618–3624 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ngamukote S., Yanagisawa M., Ariga T., Ando S., and Yu R. K. (2007) Developmental changes of glycosphingolipids and expression of glycogenes in mouse brains. J. Neurochem. 103, 2327–2341 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 39. Denslow S. A., and Wade P. A. (2007) The human Mi-2/NuRD complex and gene regulation. Oncogene 26, 5433–5438 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 40. Hassig C. A., Tong J. K., Fleischer T. C., Owa T., Grable P. G., Ayer D. E., and Schreiber S. L. (1998) A role for histone deacetylase activity in HDAC1-mediated transcriptional repression. Proc. Natl. Acad. Sci. U.S.A. 95, 3519–3524 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kou X.-X., Hao T., Meng Z., Zhou Y.-H., and Gan Y.-H. (2013) Acetylated Sp1 inhibits PTEN expression through binding to PTEN core promoter and recruitment of HDAC1 and promotes cancer cell migration and invasion. Carcinogenesis 34, 58–67 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 42. Cai B. H., Hsu P. C., Hsin I. L., Chao C. F., Lu M. H., Lin H. C., Chiou S. H., Tao P. L., and Chen J. Y. (2012) p53 acts as a co-repressor to regulate keratin 14 expression during epidermal cell differentiation. PLoS One 7, e41742 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Xiong Z., and Laird P. W. (1997) COBRA: A sensitive and quantitative DNA methylation assay. Nucleic Acids Res. 25, 2532–2534 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.