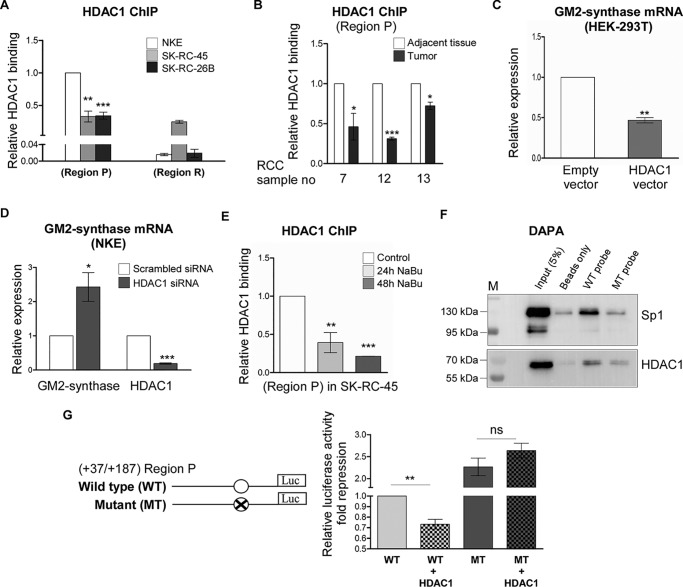
Figure 7.
Binding of Sp1 at Region P recruits HDAC1 to repress GM2-synthase transcription. A, HDAC1 associates more with transcriptionally repressed GM2-synthase gene in NKE. ChIP assay followed by qPCR for HDAC1 binding at Region P was carried out in the mentioned cell lines. Region R binding was used as negative control. The data represent three independent determinations (average ± S.E.; Student's t test; **, p < 0.01; ***, p < 0.001 versus NKE). B, HDAC1 associates more with transcriptionally repressed GM2-synthase gene in human RCC tumor-adjacent tissues. ChIP assay followed by qPCR for HDAC1 binding at Region P was carried out in the human RCC tumor and corresponding tumor-adjacent tissues. The results are expressed as relative -fold change with respect to Region P of corresponding tumor-adjacent tissues. Error bars represent mean ± S.E. of three independent determinations; Student's t test; *, p < 0.05; ***, p < 0.001. C, GM2-synthase mRNA expression is down-regulated in HEK-293T upon transient over-expression of HDAC1. HEK-293T cells were transiently transfected with HDAC1 or empty plasmids as control. 60 h post transfection, total RNA was isolated for reverse transcription. cDNAs were subjected to qPCR using GM2-synthase primer. Relative expression values were normalized to the GAPDH transcripts levels and represented as -fold change with respect to control. The data represent three independent determinations (average ± S.E.; Student's t test; **, p < 0.01). D, HDAC1 knock down increases GM2-synthase transcription in NKE cells. NKE cells were transfected with siRNA directed against HDAC1 mRNA or scrambled siRNA oligo as control. GM2-synthase was qPCR amplified as mentioned in C. The data represent four independent determinations (average ± S.E.; Student's t test; *, p < 0.05; ***, p < 0.001). E, butyrate treatment reduces HDAC1 association at Region P of GM2-synthase gene. ChIP assay performed with SK-RC-45 treated with 1 mm NaBu at mentioned time points using antibody specific for HDAC1. Precipitated chromatin DNA was estimated by qPCR. Error bars represent mean ± S.E. of three independent determinations for Region P (average ± S.E.; Student's t test; **, p < 0.01; ***, p < 0.001). F, Sp1 recruits HDAC1 to the Sp1-binding site of Region P. Nuclear extract of HEK-293T cells transfected with WT-Sp1 and HDAC1 plasmids was analyzed by DAPA, using 22-bp biotinylated probes designed from the Region P of GM2-synthase gene encompassing Sp1-binding site (Sp1 WT probe) and corresponding Sp1 site mutated oligo (Sp1 MT probe). Proteins binding to the biotinylated probes were pulled down by streptavidin beads and probed for Sp1 and HDAC1 proteins with indicated antibodies by immunoblotting. Beads only panel was included as a negative control. G, HDAC1 fails to repress luciferase activity of Region P construct encompassing mutated Sp1-binding site. Map of luciferase constructs showing WT- and MT-Sp1–binding site encompassing the Region P of GM2-synthase gene. HEK-293T cells were transiently co-transfected with WT-Sp1 or MT-Sp1 luciferase constructs along with HDAC1 or empty plasmids as control. Protein lysates were prepared for luciferase assay. The data were expressed as -fold induction relative to HEK-293T cells co-transfected with empty vector and WT-Sp1 construct and represented from five independent determinations (average ± S.E.; Student's t test; ns, not significant; **, p < 0.001).