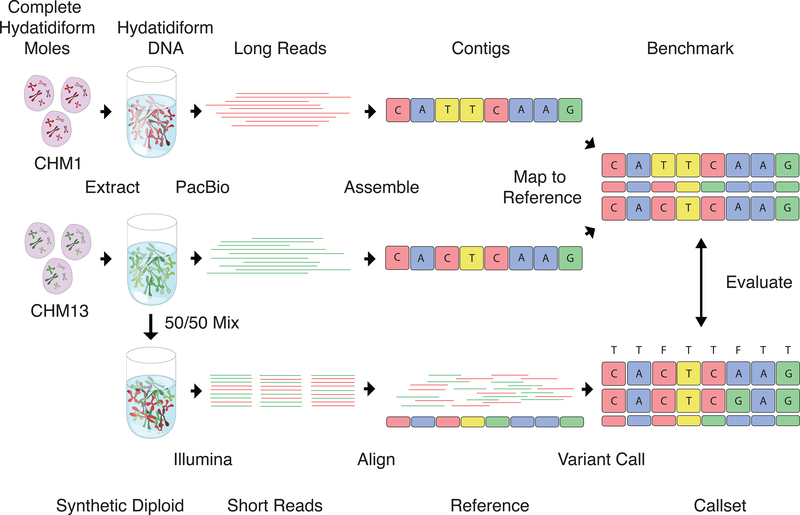
Fig. 1.
Constructing the Syndip benchmark dataset. CHM1 and CHM13 cell lines were sequenced with PacBio and de novo assembled independently. Assembly contigs were aligned to the human reference genome. Differences in the alignment were taken as ‘true’ SNPs and INDELs; regions covered by exactly one contig from each CHM assembly were identified as confident regions where true variants can be called to high accuracy. For the evaluation of diploid variant calling with short reads, equal quantities of DNA from the two cell lines were experimentally mixed. A PCR-free library was constructed from the mix and sequenced to ~45-fold coverage with 151bp paired-end reads. Variants called from the short reads were compared to the PacBio variants as truth to measure variant caller accuracy.