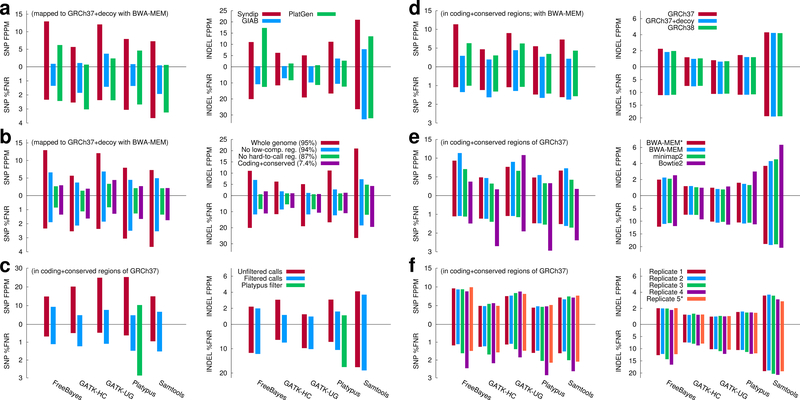
Fig. 2.
Evaluating variant calling accuracy with Syndip. %FNR denotes percent false negative rate, and FPPM is the number of false positives per million bases. (a) Comparison of Syndip, GIAB and PlatGen benchmark datasets on filtered calls. For GIAB and PlatGen, variants were called from the HiSeq X Ten run ‘NA12878_L7_S7’ available from the Illumina BaseSpace. (b) Effect of evaluation regions. Low-complexity regions were identified with the symmetric DUST algorithm. The ‘hard-to-call’ regions include low-complexity regions, regions unmappable with 75bp single-end reads and regions susceptible to common copy number variations. Panels (c)–(f) only show metrics in ‘coding+conserved’ regions. (c) Effect of variant filters. Green bars applied Platypus built-in filters. (d) Effect of the human genome reference build. Decoy sequences17 are real human sequences that are missing from GRCh37. (e) Effect of the mapping algorithms and post-processing. BWA-MEM* represents alignment post-processed with base quality recalibration and INDEL realignment; other alignments were not processed with these steps. (f) Effect of replicates. Replicate 1–4 were sequenced from four independent libraries, respectively, by mixing equal amount of DNA prior to library construction. Replicate 5* was generated by computationally subsampling and mixing reads sequenced from the two CHM cell lines separately. Replicate 1 is used in panels (a)–(e). Numerical data and the script to generate the figure are available as Supplementary Data.