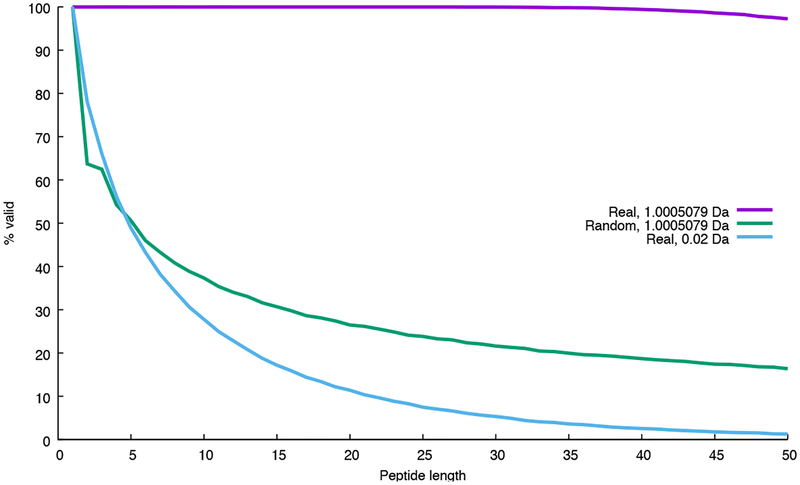
Figure 1:
Why dynamic programming cannot be performed using high-resolution mass bins. The figure plots, as a function of peptide length, the proportion of randomly generated peptide sequences that obey Equation 1. The two series marked “Real” use monoisotopic masses from the 20 real amino acids; the series marked “Random” uses masses that have a random number in the range 〈0, 1] added to each mass. Two bin sizes (1.005079 Da and 0.02 Da) are used. For each series, a total of 100,000 peptides were simulated. With small bin sizes, the large proportion of peptide sequences whose masses violate Equation 1 causes the dynamic programming to fail.