Table 2.
Zymoseptoria tritici core biotrophic effector candidate genes
| Criteria | Significantly up‐regulated at stage B | |||
| Low/no expression during stage C and D | ||||
| RPKM stage B > RPKM stage C in Zt05 and Zt09 | ||||
| Expression profiles |
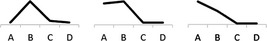
|
|||
| Putative functions | Bypass host recognition during biotrophic colonization | |||
| Candidate genes | 25 (Supporting Information Figure S13) | |||
| Candidates encoding hypothetical proteins | 23 (Supporting Information Table S15) | |||
| Candidates with predicted function | Gene | Function/Functional prediction | PFAM domains | GO terms associated |
| MgNLP (Motteram et al., 2009) Zt09_chr_13_00229 | Necrosis and ethylene‐inducing peptide 1‐like protein MgNLP (Kettles, Bayon, Canning, Rudd, & Kanyuka, 2017; Motteram et al., 2009) | PF05630 | GO:0008150; GO:0003674; GO:0005575 | |
| Zt09_chr_2_00129 | Hypothetical protein, secreted phospholipase A2 precursor | PF06951 | GO:0004623; GO:0005509; GO:0005576; GO:0016042 | |
Summary of core Z. tritici biotrophic effector candidate genes that were identified based on their specific expression profiles within the Z. tritici core transcriptional program during wheat infection. Functional annotation, PFAM, and GO term information from Grandaubert et al. (2015).
