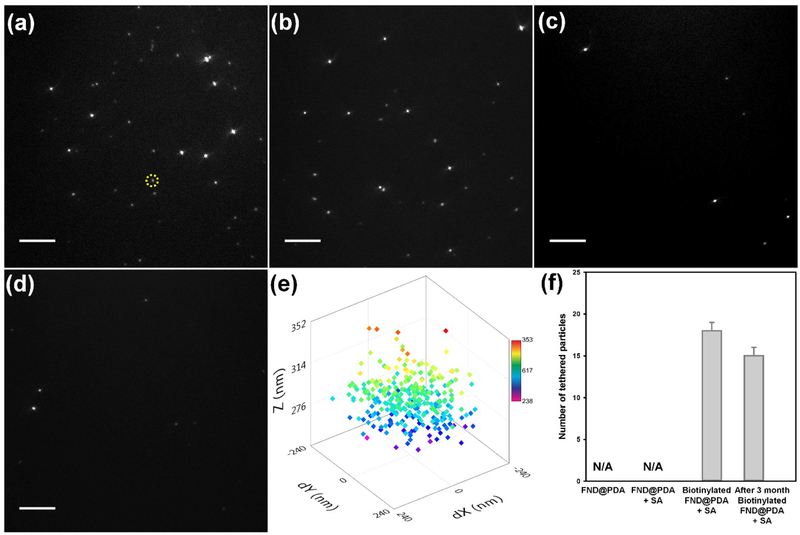
Figure 6.
DNA tethering experiments under four different conditions. TIRFM images for DNA tethering with (a) biotinylated FND@PDA treated with SA (FND@PDA-PEG-biotin+SA), (b) FND@PDA-PEG-biotin stored for three months at RT in water, treated with SA, (c) PDA-coated FND (FND@PDA), and (d) FND@PDA treated with SA (FND@PDA+SA). Scale bar in panels a-d is 10 μm. (e) An example 3-D spatial distribution of a DNA tethered FND@PDA-PEG-biotin+SA obtained by tracking the Brownian motion of the particle (indicated by the yellow circle in panel a). Color scale bar provides a measure of the z position (in nm) of the tethered FND. (f) The average number of DNA molecules tethered by FND per a field of view for four different conditions. No DNA tethered FND particles were observed for either FND@PDA or FND@PDA+SA. A few nonspecifically stuck immobile particles are distinguished from tethered particles which undergo Brownian motion, (please see the video S3 and S4). The average number of FNDs tethered per field of view (73 μm × 73 μm) with the day-old and 3-month old FND@PDA-PEG-biotin+SA are 18 ± 1 [N (number of different field of views) = 6)] and 15 ± 1 [N = 8)] respectively. The reported error is the standard error of the mean.