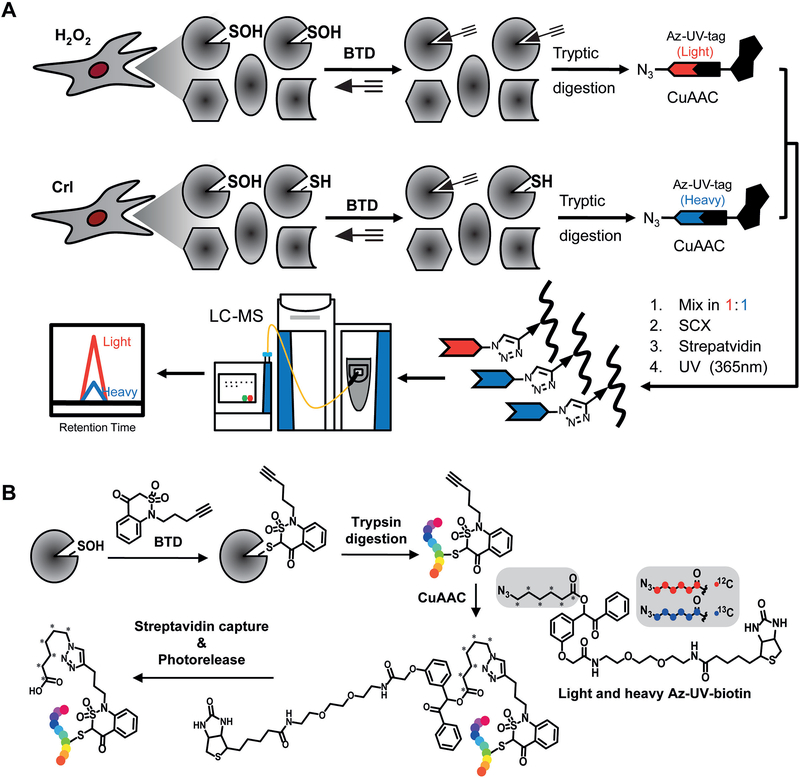
Figure 1. BTD-based quantitative S-sulfenylome analysis.
(A) Workflow for proteome-wide analysis of protein S-sulfenylation in RKO cells. S-sulfenylated proteins in cells treated with or without H2O2 are labeled with the BTD probe in vitro or in situ. Cell proteins then were digested with trypsin and labeled peptides are further conjugated with light (H2O2 treated) and heavy (vehicle control) azido biotin with a photocleavable linker, cleaned with SCX spin columns, captured with streptavidin beads, and photoreleased. The resulting peptides were analyzed by LC-MS/MS. (B) Chemical transformation of BTD-labeled form in the entire chemoproteomic workflow.