Figure 1.
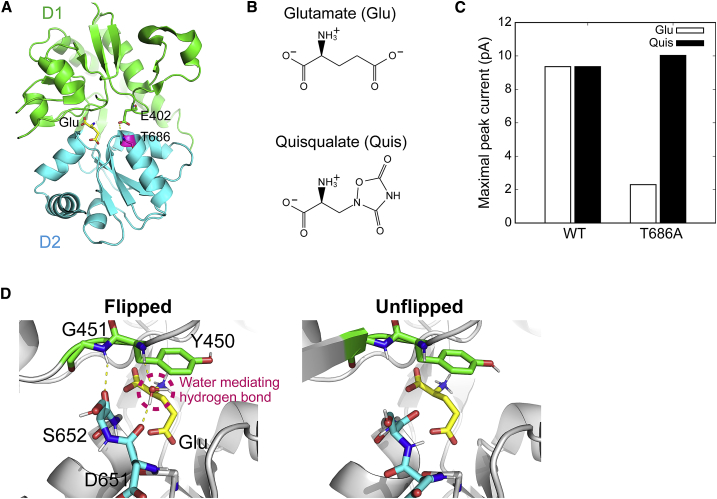
Structure of ligand-binding domain (LBD) and effect of T686A mutation on agonist efficacy. (A) A cartoon representation of the LBD of GluA2 with glutamate (Glu). The bound Glu, E402, and T686 are shown in stick representation. The D1 and D2 lobes are colored green and cyan, respectively. The glutamate and T686 are colored yellow and magenta, respectively. A yellow dashed line represents hydrogen-bonding interaction. (B) Chemical structures of Glu and Quis. (C) Maximal peak currents induced by channel opening upon ligand binding. These data are taken from the experimental results of Zhang et al. (19). (D) Flipped and unflipped conformations of the peptide backbone of D651 and S652. Yellow dashed lines represent hydrogen-bonding interactions. To see this figure in color, go online.