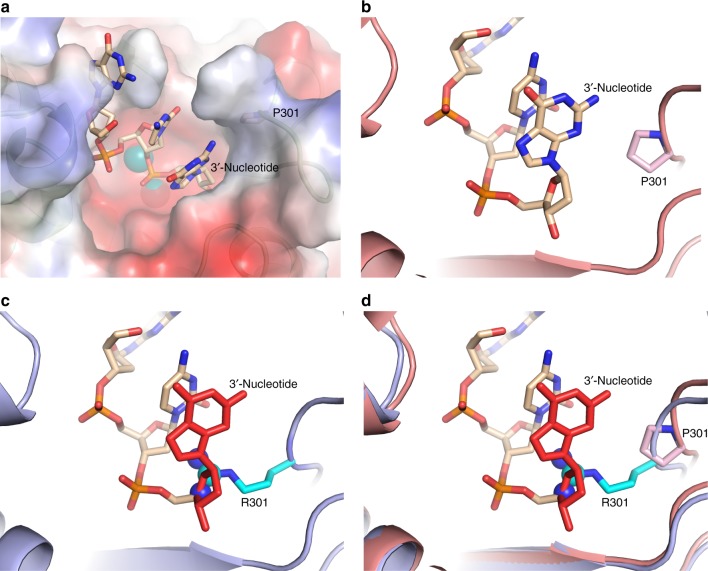
Fig. 4.
Modelling DNA into the exonuclease site of Pol2CORE. a The electrostatic surface view of the wild-type exonuclease domain of Pol2CORE with the modeled position of single stranded DNA (ssDNA) in the exonuclease active site. A representative structure of the ssDNA from our molecular dynamics simulations is shown as sticks (wheat color). b A representative structure of DNA in the wild-type exonuclease domain of Pol2CORE from the molecular dynamics simulation in cartoon view. The 3´-end of the ssDNA is shown in the front with P301 (in light pink) in the exo-loop on the right hand side. c The crystal structure of P301R overlayed with the ssDNA from (b). The terminal nucleotide (in red) occupies the same space as R301 (the atoms in the guanidine group shown as spheres). d The crystal structure of P301R overlayed with the best representative model from molecular dynamics simulations with wild-type exonuclease domain and ssDNA from (b). The terminal nucleotide (in red) occupies the same space as R301 (with the guanidine group shown as spheres)