Fig. 8.
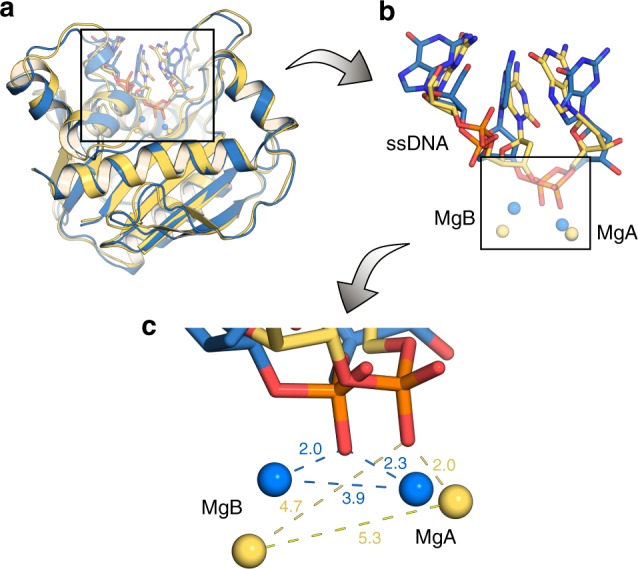
Comparisons of the exonuclease active site of wild-type and P301R Pol2CORE after molecular dynamics simulations. Overlay of structures of the exonuclease active site of wild-type (blue) and P301R Pol2CORE (yellow), taken as representative snapshots from 3 × 200 ns simulations of each system, and zooming in progressively from (a) the full tertiary structure to (b) the exonuclease-site to (c) the metal binding site. As can be seen, the proposed bridging conformation of the phosphate group from the ssDNA backbone is lost in the P301R variant, due to displacement of the Mg2+ ion in the B-site. The distances annotated in (c) are average distances (Å) over the entire simulation and all replicas
