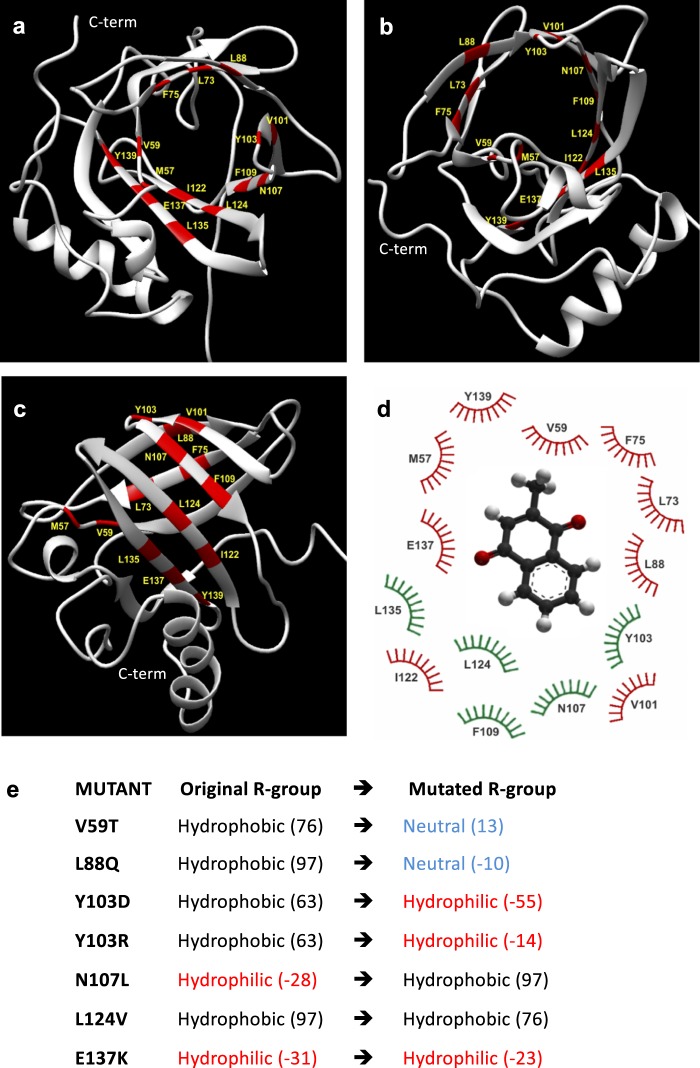
Figure 2.
(a–c) The structure of wild type MUP20 (PDB ID: 2L9C, 6xHis tag is not represented) in different orientations. The 3D representation shows in red the amino acids belonging to the hydrophobic pocket involved in the binding process, according to the LPC/CSU analysis. C-term indicates C-terminus. (d) Relevant residues involved in the ligand-MUP20 interaction. Those in green are conserved across the natural MUP isoforms, while the variable residues are in red. (e) Amino acid mutations in MUP20. The polar/apolar nature of the original and substituted amino acids is reported together with the corresponding hydrophobicity index values (in brackets)36.