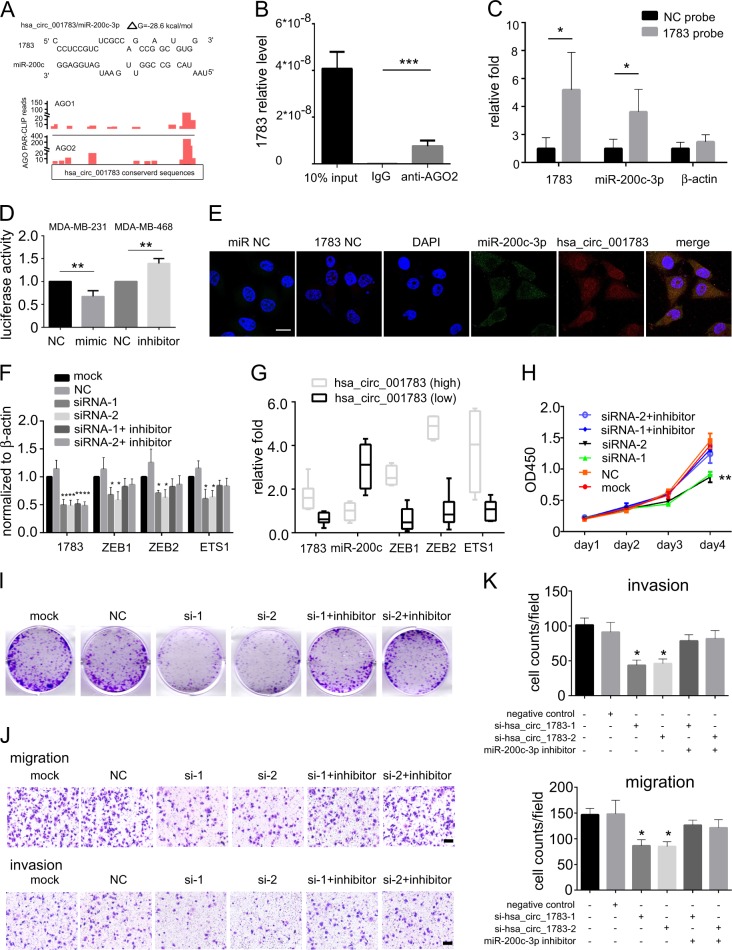
Fig. 6. Hsa_circ_001783 serves as sponge for miR-200c-3p.
a Examples of the potential bindings between hsa_circ_001783 conserved sequence and miR-200c-3p. Hsa_circ_001783 conserved locus is densely bound by AGO1 (red) and AGO2 (red). b AGO2 RNA-binding protein immunoprecipitation. 1783 is referred as hsa_circ_001783. All data are shown as the mean ± SD. ***P < 0.001 compared to IgG. c circRNA pull-down assay. 1783 is referred as hsa_circ_001783. **P < 0.01 compared to negative control (NC) probe. d Luciferase assay of MDA-MB-231 and MDA-MB-468 co-transfected with luciferase reporter containing hsa_circ_001783 conserved sequences and miR-200c-3p mimic or miR-200c-3p inhibitor. NC represents mimic negative control and inhibitor negative control. All data are shown as the mean ± SD. **P < 0.01 compared to negative control. e RNA fluorescence in situ hybridization for co-localization of hsa_circ_001783 and miR-200c-3p in MDA-MB-231. Scale bar 10 μm. f The expression of hsa_circ_001783 and miR-200c-3p mRNA targets: ZEB1, ZEB2, and ETS1 after hsa_circ_001783 knockdown. g qPCR analysis of the expression of hsa_circ_001783, miR-200c-3p, and miR-200c-3p mRNA targets: ZEB1, ZEB2, and ETS1 in breast cancer clinical specimens. h The proliferation status of MDA-MB-231 after siRNA transfection or miR-200c-3p inhibitor co-transfection determined by CCK-8. OD optical density. All data are shown as the mean ± SD; **P < 0.01 compared to mock. i Colony formation ability of MDA-MB-231 after siRNA transfection or miR-200c-3p inhibitor co-transfection. j The representative images of migrated and invaded MDA-MB-231 after siRNA transfection or miR-200c-3p inhibitor co-transfection. Scale bar, 100 μm. k The migration and invasion abilities of MDA-MB-231 after hsa_circ_001783 knockdown or miR-200c-3p inhibitor co-transfection. All the data are shown as the mean ± SD; *P < 0.05, compared to mock