Abstract
Introduction:
We aimed to determine the prevalence and landscape of germline mutations among patients with young onset pancreatic ductal adenocarcinoma (PDAC) as well as their influence in prognosis.
Methods:
Patients from two cohorts were studied, the High Risk Cohort (HRC) which included 584 PDAC patients who received genetic counseling at MD Anderson Cancer Center and a General Cohort (GC) with 233 metastatic PDAC patients. We defined germline DNA sequencing on 13 known pancreatic cancer susceptibility genes. The prevalence and landscape of mutations was determined and clinical characteristics including survival were analyzed.
Results:
A total of 409 patients underwent genetic testing (277 from HRC and 132 from GC). As expected, the HRC had higher prevalence of germline mutations compared to the GC: 17.3% vs 6.81%. The most common mutations in both cohorts were in BRCA1/2 and mismatch repair (MMR) genes. Patients younger than 60 years old had significantly higher prevalence of germline mutations in both the HRC (OR: 1.93 +/−1.03–3.70, P: 0.039) and GC (4.78 +/−1.10–32.95, P: 0.036). Furthermore, PDAC patients with germline mutations in the GC had better overall survival than patients without mutations (HR= 0.44, 95% CI of HR: 0.25–0.76, p: 0.030).
Discussion
Germline mutations are highly prevalent in patients with PDAC of early-onset and can be predictive of better outcomes. Considering emerging screening strategies for relatives carrying susceptibility genes as well as impact on therapy choices, genetic counseling and testing should be encouraged in PDAC patients, particularly those of young onset.
Keywords: Hereditary Pancreatic Cancer, Cancer Genetics, Genetic Testing, BRCA1/2, Cancer Risk Assessment
Introduction
Pancreatic ductal adenocarcinoma (PDAC) is currently the eleventh-most common cancer in incidence and the third-leading cause of cancer-related deaths in the United States (1). The majority of PDACs are sporadic; however 5–10% may have a hereditary cause (2). PDAC is considered a disease of the elderly and most diagnoses are made in individuals over the age of 65, with a median age at diagnosis of 70 years (3). Individuals diagnosed with PDAC under the age of 60 are considered to be young-onset and potentially at high risk for a genetic predisposition. While pancreatic cancer is typically associated with environmental and lifestyle factors such as smoking, obesity and diabetes (4–7), inherited germline mutations confer a significantly elevated lifetime risk for PDAC. Germline mutations in a growing number of genes have been associated with increased risk of PDAC, including ATM, APC, BRCA1, BRCA2, CDKN2A, MLH1, MSH2, MSH6, PMS2, PALB2, STK11, and PRSS1 (2, 8–11).
Familial pancreatic cancer (FPC) is defined as a family with at least two first-degree relatives with PC without an identifiable syndrome or genetic mutation in the family. Relatives meeting FPC criteria have an empirically increased risk to develop PDAC over the general population; these individuals can be even further stratified depending on their degree of relationship to the affected relative(s) (12). The standard incidence rates (SIR) to develop PDAC in individuals with one first-degree relative, two first-degree relatives, or three or more first-degree relatives are 3–4, 5–7, and 17–32, respectively (3, 13).
Germline mutations confer an elevated lifetime risk for PDAC (2, 8–11). Typically germline mutations associated with higher risk for PDAC have been suggested by kindreds with multiple generations of pancreatic or related cancers (breast, ovarian, colon, etc.), early cancer diagnoses, individuals with multiple primary tumors, and/or Ashkenazi Jewish ethnicity(13).
Recent studies have found that an appreciable fraction of patients with apparent sporadic pancreatic cancers have germline mutations (14, 15). In search of predictive factors for germline mutations, previous studies have looked at age of presentation but results have been ambiguous. Grant et al (16) and Holter et al (17) reported germline mutations prevalence in PDAC patients of 3.8% and 4.5% respectively, while Hu et al(18) reported 9.4% prevalence of mutations in established PDAC genes and up to 13.5% total pathogenic mutations. These three studies found no differences in age with respect to mutations but the numbers of young onset PDAC included in the analysis was rather low. Very recently, a major study from Shindo et al (15) has found slightly higher prevalence of germline mutations in younger patients. Therefore we aimed to definitively determine if patients with germline mutations present earlier than those without mutations. Given that only 30% of individuals with PDAC are diagnosed under the age of 60 (ref17), we examined germline mutation prevalence in a high risk cohort which includes a large young-onset PDAC subpopulation during an 11-year period at MDACC. As validation, we examined a general cohort of PDAC patients also seen at MDACC.
Materials and Methods
Patient Selection
High-Risk Cohort (HRC):
Included patients referred to genetic counseling, based on established criteria (Supplementary Table 1), seen at the University of Texas MD Anderson Cancer Center (MDACC) from 2005 to 2016. All patients diagnosed with PDAC diagnosed under the age of 60 (young onset PDAC) met referral criteria regardless of family history. From a total 584 patients seen in the HRC, 261 were patients with young onset PDAC. The standard genetic counseling consultation included obtaining a pedigree of at least three generations, risk assessment, and review of the patient’s personal medical history and risk factors. Genetic testing was recommended based on formal risk assessment made by a board-certified genetic counselor (S.A.B. and M.E.M.). All genetic testing was performed at Clinical Laboratory Improvement Amendments-certified laboratories. The landscape of genetic testing options has dramatically evolved over the time during which these patients were seen for genetic counseling. Single-gene analysis based on personal or family history or multi-gene panel testing were performed. In the panels up to thirteen mutations were tested: ATM, APC, BRCA1, BRCA2, CDKN2A, MLH1, MSH2, MSH6, PMS2, PALB2, STK11, TP53, and EPCAM.
General Cohort (GC):
All patients with metastatic pancreatic cancer that received first line chemotherapy at MDACC, between January 2010 and January 2016, and consented for DNA testing, were eligible. Results were considered for research purposes only and not used to make clinical decisions. Exome capture was performed from 500 ng of genomic DNA using the KAPA library (KAPA Biosystems, Willmington, MA) and sequenced using an Illumina HiSeq 2500 instrument with 200X average coverage. Genetic variants were classified based on available public databases (Exome Aggregation Consortium (EXAC) Database 0.3, COSMIC release 70,72, NHLBI exome sequencing project ESP6500SI-V2, dbSNP129,138, ClinVar v20150330, Exome Variant Server (EVS), 1000 Genomes, dbVar, Human Gene Mutation Database, HGVS, and DECIPHER) accessed between 01/2017 and 12/2017 and in-silico analysis (SIFT, PolyPhen-2, MutationTaster). For the present study we have focused in analyzing the 13 same genes that were included in the multi-gene panel testing performed in HRC patients.
For both cohorts, clinical and pathological data were abstracted from the Electronic Health Record. MDACC Institutional Review Board (IRB) approved this study. Study was conducted in accordance with Belmont report.
Only patients diagnosed with a pathogenic germline mutation (in both cohorts) were assigned to the hereditary group and those with a variant of uncertain significance (VUS) or no identifiable mutations were assigned to the sporadic group. Patients harboring a pathogenic gene mutation and an additional variant of uncertain significance (VUS) were assigned to the hereditary group.
Statistical Analysis
Categorical variables are reported as frequencies and percentages; continuous data are summarized as mean and standard deviation (SD). Chi-squared and Fisher’s exact tests were used to evaluate associations between categorical variables and mutation status. The t-test was used to compare the distributions of continuous variables (such as age) between mutation statuses. Univariate logistic regression model were used to evaluate the association between the risk factor (age) and mutation status. Odds ratios (OR) and 95% confidence intervals (CI) were estimated to measure the strength of association. The recursive partitioning method was used to select optimal cut-off point for continuous age based on mutation status using HRC. The identified optimal cut-off point for age was validated using GC. Overall survival (OS) was defined as the time from diagnosis to death from any cause. Living patients were censored at date of last follow-up. Kaplan-Meier curves were estimated for the survival distributions by mutation status. The Log-rank test was used to test the difference in survival distributions between subgroups. Univariate Cox proportional hazard models were used to determine the effects of mutation status on OS. Hazard ratios and 95% confidence intervals were provided. All tests are two-sided. P-values less than 0.05 are considered statistically significant. All analyses were conducted using SAS 9.4 (SAS, Cary, NC) and S-Plus 8.0 (TIBCO Software Inc., Palo Alto, CA) software.
Results
Patients
High Risk Cohort
Between 2005 and 2016 five hundred and eighty-four patients with PDAC received genetic counseling. The characteristics of this population are described in Table 1. The mean age at diagnosis was 61.44 years and slightly more than half were female (53.8%). In terms of racial/ethnicity characteristics, the majority of patients were non-Hispanic white (82.7%), while Hispanic, Black, and Asian represented 6.8%, 7.2% and 3% respectively. A large proportion of patients had no previous history of tobacco use (58.2%) while alcohol consumption was reported by 62.7% of patients. Importantly, 13.6% of patients had personal history of breast cancer and 4.5% had gynecological cancer. A total of 171 patients were found to have at least one first-degree relative (FDR) with breast cancer (29.2%). With respect to family history of pancreatic cancer, 127 patients had at least one first-degree relative (21.7%) and 111 had at least one second-degree relative (SDR) affected (19%).
Table 1.
Patient demographics, personal and family History in HRC (n=584). FDR, first-degree relative; SDR, second-degree relative. Age is shown in years with range.
| Characteristic | High Risk Cohort (n=584) |
|---|---|
| Age at diagnosis (Mean) | 61.44 (25–89) |
|
Sex Female Male |
310 (53.1%) 274 (46.9%) |
|
Race White Hispanic Black Asian Unknown |
183 (82.7%) 40 (6.8%) 42 (7.2%) 18 (3.0%) 1 (0.17%) |
|
Smoking History Never Past/Current |
340 (58.2%) 244 (41.8%) |
|
Alcohol History Never Occasional/Heavy |
218 (37.3%) 366 (62.7%) |
| History of Pancreatitis | 54 (9.2%) |
| Chronic Diabetes | 96 (16.4) |
| New-Onset Diabetes | 79 (13.5%) |
|
Personal History of Cancer Breast Gynecologic Melanoma Colon |
80 (13.6%) 26 (4.45%) 13 (2.2%) 25 (4.2%) |
|
Family History of Cancer ≥1 affected FDR Breast Pancreas ≥1 affected SDR Breast Pancreas |
171 (29.2%) 127 (21.7%) 166 (28.4%) 111 (19.0%) |
From the 584 patients in the HRC, 277 underwent genetic testing for hereditary pancreatic cancer genes. We compared the clinical and pathological characteristics of patients who underwent testing versus those who did not and we found that younger patients were more likely to be tested (p=0.019). We then observed that patients with personal history of chronic pancreatitis were less likely to be tested (p=0.014) and those with a personal history of breast or gynecological cancers were more likely to undergo genetic testing (p=0.0001 and p=0.0085). Those with at least one first-degree relative with breast cancer were also more likely to undergo genetic testing than those without a first-degree relative with breast cancer (p=0.0047) (Supplementary Table 2). These results were expected as young patients, or those with stronger personal or family history of cancer received a stronger recommendation for genetic testing at time of genetic counseling.
Outcomes of Genetic Counseling and Testing in HRC
Pathogenic germline mutations were identified in 48 patients (17.32%) (Fig.1A), VUS were found on 14 patients, and 215 patients had uninformative results. Of the 277 patients who underwent clinical genetic testing in the HRC, 240 (86.7%) were tested by single-gene analysis and 37 (23.3%) by multigene panel. Of the 130 young-onset (<60) PDAC patients, 28 (21.6%) underwent panel testing and 102 (78.4%) had single-gene testing. In the young-onset cohort, we compared the mutation detection of panel versus single gene testing. We found no significant difference in the yield of pathogenic mutations detected with panel versus single gene testing, 21.43% versus 22.5%, respectively (p=0.9). However, panel testing identified a significantly higher number of VUS than single gene testing, as expected (25% versus 1.96%; p=0.0003) (Supplementary Fig. 1). There was significant heterogeneity in the number of patients tested for each gene, as single-gene(s) genetic testing was determined by a genetic counselor based on personal and family history risk assessment; therefore, not all patients in the HRC were tested for all genes. The most frequently mutated genes were BRCA1 and BRCA2 (29/128 patients with BRCA2 mutations and 5/127 with BRCA1, respectively) which is consistent with previous reports (17). The second most common group of mutations detected were in the MMR genes with 2/33 in MLH1, 3/34 in MSH2 and 2/33 in MSH6. Seven patients had germline mutations in other genes: 2/35 in TP53 and ATM (30 tested) and 1 each in APC (31 tested), CDKN2A (37 tested) and STK11 (33 tested) (Supplementary Fig.2A). Variants of uncertain clinical significance (VUS) were considered as negative due to inconclusive association with PDAC predisposition. There were three patients who carried both one pathogenic mutation and one VUS. For analysis purposes, these patients were considered in the group of patients with pathogenic mutations. Details on the pathogenic mutations found in HRC young onset patients are listed in Supplementary Table 3.
Figure 1. A. Flow Diagram.
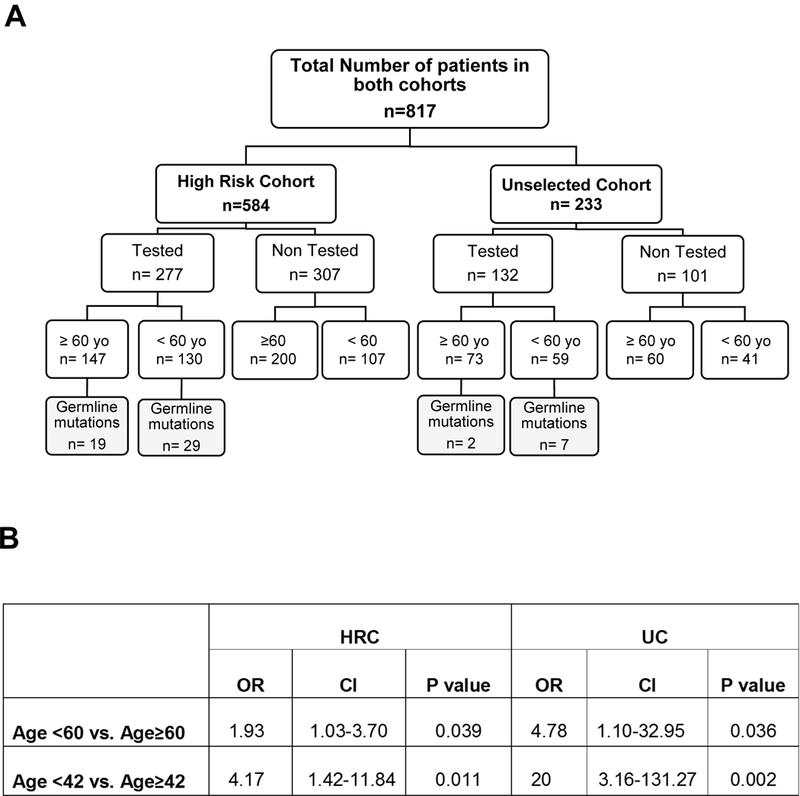
Mutation status by age of diagnosis in both cohorts (HRC and GC); B. Statistical analysis assessing the association between mutation status and age (age ≥ 60 years vs < 60 years and age ≥ 42 years vs < 42 years) in HRC and GC. OR: Odds Ratio; CI: Confidence Interval.
Influence of age in HRC genetic testing results
We then looked at the influence of age in the mutation status of tested patients within the HRC, which was the main goal of the study. We found that the mean age at time of PDAC diagnosis in patients with germline mutations was significantly younger than in patients without mutations (56.6 (SD+/−10.95) vs. 61.07 (SD+/− 10.95), (P=0.010, Table 2). We then compared mutation prevalence in early-onset PDAC patients (<60 years-old) vs. older patients (≥ 60 years-old) and found that early-onset patients had significantly higher odds of testing positive compared with older patients (OR 1.93, 95%CI: 1.03–3.70, P=0.039) (Fig. 1A,B).
Table 2.
Comparison of Demographical Information, Clinical Characteristics, and Family History of Cancer in patients from the HRC who tested positive vs. negative for mutations. Variants of Uncertain Significance (VUS) were tabulated as negative results. FDR, first-degree relative; SDR, second-degree relative.
| Characteristic | Mutation positive (n=48) n (%) |
Mutation negative (n=229) n (%) |
p-value |
|---|---|---|---|
| Age | |||
| Mean (yrs +/− SD) | 56.6 (+/−10.95) | 61.07 (+/−10.95) | 0.010* |
| Sex | |||
| Female Male |
22 (45.8) 25 (54.2) |
127 (55.5) 102 (44.5) |
0.2223 |
| Race | |||
| White Hispanic Asian Black |
35 (72.9) 8 (16.6) 2 (4.1) 3 (6.2) |
200 (87.3) 11 (4.8) 11 (4.8) 7 (3.0) |
0.026* |
| Stage | |||
| Localized Borderline Metastatic |
16 (33.3) 7 (14.6) 25 (52.1) |
68 (29.7) 33 (14.4) 128 (55.9) |
0.870 |
| Grade of Differentiation | |||
| Well Moderately Poorly |
0 (0) 18 (58.1) 13 (41.9) |
2 (1.3) 115 (72.3) 42 (26.4) |
0.220 |
| Smoking History | |||
| Current Past Never |
8 (16.7) 11 (22.9) 29 (60.4) |
43 (18.8) 42 (18.3) 144 (62.9) |
0.752 |
| Alcohol History | |||
| Heavy Occasional Never |
1 (2.1) 26 (54.2) 21 (43.8) |
14 (16.1) 142 (62) 73 (31.9) |
0.244 |
| History of Pancreatitis | 1 (2.1) | 16 (7) | 0.322 |
| Chronic Diabetes | 10 (20.8) | 36 (15.7) | 0.386 |
| Personal History of Cancer | |||
| Breast Gynecologic Melanoma Colon |
13 (27.1) 2 (4.2) 2 (4.2) 3 (6.3) |
43 (18.8) 17 (7.4) 7 (3.1) 11 (4.8) |
0.192 0.543 0.657 0.715 |
| Family History of Cancer | |||
|
≥1 affected FDR Breast Gynecologic Melanoma Pancreas Colon ≥1 affected SDR Breast Gynecologic Melanoma Pancreas Colon |
23 (56.1) 10 (34.5) 5 (10.4) 11 (23.4) 12 (25) 14 (29.2) 9 (18.8) 1 (2.1) 7 (14.9) 7 (18.9) |
74 (36.8) 29 (17.3) 16 (7) 52 (22.7) 28 (12.2) 72 (31.4) 18 (7.9) 11 (4.8) 50 (21.8) 25 (13.4) |
0.121* 0.031* 0.379 0.917 0.022* 0.756 0.020* 0.698 0.284 0.385 |
Using recursive-partitioning method, we identified 42 years of age as the optimal cut-off that best separates age-stratified groups based on mutation status in the HRC. Patients younger than 42 years old had remarkably higher odds of testing positive for a mutation than patients older than 42 years (OR: 4.17, 95%CI:1.42–11.84, P= 0.011) (Fig.1B).
Importantly, we found that approximately half (50.1%) of the patients in the HRC met National Comprehensive Cancer Network (NCCN©) 2018 BRCA1/BRCA2 genetic testing criteria (Supplementary Table 4) and 44.6% of patients younger than 60 years old met criteria, indicating that 55.4% of young onset patients were tested solely based on indication of age. Finally, in the mutation positive HRC, 52.1% of patients met criteria while in the mutated younger onset group 41.4% did (Supplementary Table 5).
Comparison of mutation-positive versus mutation-negative patients in HRC
We then assessed for the association between other potential predictive factors and mutational status and a significant difference was seen by ethnicity with Hispanic individuals more likely to test positive (P=0.026) (Table 2). With respect to family history, patients with at least one or more first-degree relatives (FDR) with breast, gynecological and colon cancer were significantly more likely to harbor mutations (P= 0.021, P= 0.031, P= 0.022 respectively) (Table 2). Also, patients with at least one second-degree relative (SDR) with gynecological cancers were more likely to have germline mutations. Patients with family history contributory for pancreatic cancer in FDRs and/or SDRs were not more likely to have germline mutations than those without it (Table 2). There were no other significant differences between the two groups (with mutations vs. without mutations) regarding sex, grade of differentiation, stage, personal history of cancer, and other risk factors.
General Cohort
To validate the higher prevalence of germline mutations in early-onset PDAC in a non-high-risk cohort, we analyzed a second group of patients with PDAC seen at the same institution referred to as the General Cohort (UC). A total of 233 consecutive patients with metastatic PDAC receiving chemotherapy treatment at MD Anderson were enrolled from which 132 patients had sequencing performed. The selection of patients for sequencing was randomly performed. The characteristics of the tested GC population are described in Supplementary Table 6. The mean age at diagnosis was 59.73 years and 59.1% of the patients were male. Most of patients were non-Hispanic white (84%), and Hispanic, Black, and Asian represented 4.5%, 9% and 2.2% respectively. A total of 11 patients were found to have at least one FDR with PC (8.3%) while 9 patients had at least one SDR with PC (6.8%).
Genetic Testing Results in General Cohort (GC) and Influence of Age
From the 132 patients that had sequencing, 9 were found to have a pathogenic germline mutation (6.81%) (Fig.1A). Again in this group, the most common mutations found were in BRCA1/2 genes (Supplementary Fig.2B). The average age of patients with germline mutations was also younger than patients without mutations (48.44 (SD+/−9.65) vs. 60.56 (SD+/− 10.57), (P=0.033, Supplementary Table 7). In this cohort, 59 patients presented with early-onset PDAC (<60) and 7 of them had germline mutations, which is significantly associated with higher odds of testing positive for a mutation compared with older patients (>60) (OR:4.78, 95%CI:1.10–32.95, P= 0.036) (Fig.1A,B). When using the optimal cut-off of 42 years of age, determined using the HRC dataset, again we found significantly higher prevalence of mutations in patients younger than 42 vs those older (OR: 20, 95%CI: 3.16–131.27, P= 0.002) (Fig.1B). No other factors, besides age, were associated with higher prevalence of mutations in the GC (Supplementary Table 7).
Mutational status as predictive of clinical outcomes in HRC and GC
We then performed an analysis of clinical outcomes in both cohorts. We first analyzed overall survival in resectable patients from the HRC and found that patients with germline mutations had significantly better overall survival that patients without mutations, with a median survival of 70.4 months versus 32.6 months respectively (HR: 0.55, 95% CI of HR: 0.33–0.91, P= 0.03) (Fig.2A). Adjuvant chemotherapy regimens in the resectable HRC patients were comprised of single agent gemcitabine and/or platinum-based regimens including 5-fluorouracil, irinotecan, oxaliplatin (FOLFIRINOX), single-agent oxaliplatin, and cisplatin. Unfortunately, we do not have enough power to detect differences in outcomes by treatment type given the limited number of patients in each chemotherapy type.
Figure 2. Survival data.
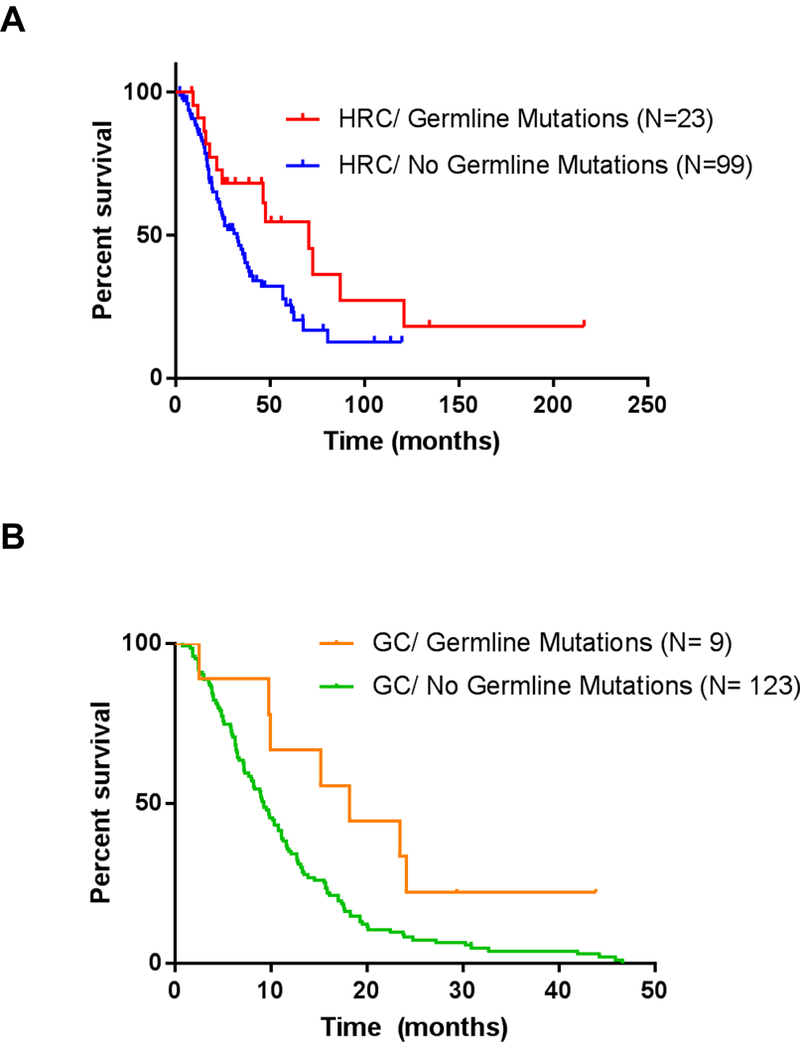
Kaplan-Meier Survival curves for resectable PDAC patients by mutation status in the HRC (A) and for metastatic patients from GC (B). N= Number of patients in each subgroup. P value calculated based on Log-rank test.
We then analyzed survival in PDAC patients from general cohort (all metastatic). Patients from the GC with germline mutations had significantly improved survival than patients without mutations detected with a median survival of 18.2 vs 9.2 months (HR: 0.44, 95% CI of HR: 0.25–0.76, P= 0.03), regardless of adjuvant chemotherapy treatment regimen (Fig.2B). When we looked at the metastatic patients from the HRC we did not find significant differences in survival based on their mutational status (Supplementary Fig.3A). However, metastatic patients in the HRC, disregarding their mutational status, had significantly longer overall survival than patients without mutations in the general cohort (HR: 0.51, 95% CI of HR: 0.39–0.66, P<0.0001) (Supplementary Fig.3B).
Discussion
This study analyzes a large HRC of patients diagnosed with pancreatic cancer at a tertiary referral center with the main goal of determining if patients with germline mutations on PDAC susceptibility genes present earlier than those with sporadic PDAC. We determined the prevalence of germline mutations and looked for predictive factors of germline mutations and used a second general cohort for validation purposes. A total of 17.32% of patients in the HRC were found to have pathogenic germline mutations versus 6.81% in the GC which is consistent with the previously reported prevalence of hereditary pancreatic cancer in the general pancreatic cancer population (15, 16). Interestingly, panel testing did not yield significantly increased detection of pathogenic germline mutations over single gene(s) testing as guided by genetic counselor risk assessment. However, panel testing did significantly increase the risk of identifying variants of uncertain significance. This finding is significant in the clinical practice context where variants of uncertain significance can pose confusion and add uncertainty to individuals attempting to discern their risk to develop PDAC. Germline mutations for genes associated with increased risk of PDAC were found in 11–22% of PDAC patients younger than 60 years old and 44–50% of patients younger than 42 years old. Therefore, young age represents a strong predictive factor of germline mutations in PDAC patients. The optimal cutoff point of 42 years old found in the High Risk cohort was exploratory and tested in the unselected cohort. However, this cutoff point should be further validated in a larger scaled study in the future.
We found that family history of breast, gynecological or colon cancer are actually predictive of germline mutations while family history of PDAC was not predictive, in agreement with previous reports (15, 16) while family history of PDAC was not more prevalent on patients with germline mutations. This data suggests that the germline mutations associated with familial PDAC may not be included in the panel of established PDAC susceptibility genes.
Regarding clinical outcomes, we have found that patients in the HRC without mutations had a shorter overall survival than those with mutations only in patients older than 60 years old. An explanation for this might be that PDAC patients younger than 60 years old who tested negative may still harbor a germline mutation in genes for which they were not tested. With respect to metastatic patients, only PDAC patients from the GC with germline mutations were found to have better clinical outcomes compared to those without mutations. Similar to the explanation above, patients from the HRC do not have substantial differences based on their mutational status because those with negative results may also have mutations, which have not been tested for.
The use of a discovery cohort with a large number of young-onset PDAC patients and a validation cohort with patients that were not at high risk for mutations based on personal and/or family history is strength of the study. A major limitation of this study is the use of panels with differing genes analyzed in individual patients of the HRC and not all patients were tested for all genes. The reason for this is that single-gene analysis of one to three genes was standard of care until 2013 when multi-gene panels became available at our institution. We compared demographic data from patients tested for individual genes or with panels and found no differences between the two groups (Supplementary Table 8) suggesting that this factor should not affect final results. Additionally, panel testing did not yield significantly higher detection of pathogenic mutations, but did significantly increase the likelihood to identify variants of uncertain significance. Future studies would ideally include comprehensive analysis of an entire cohort for the same set of genes to validate these findings. Similarly, variants identified in the GC cohort were subject to classification on the research platform involving public database review and in-silico analysis but were not subjected to the stringent clinical variant interpretation as those tested via standard clinical genetic testing in the HRC cohort.
Higher detection rates of germline mutations will potentially impact therapy choices for patients with PDAC. BRCA-associated PDAC have been shown to have higher sensitivity to platinum therapy and poly (ADP-ribose) polymerase (PARP) inhibitors(19) while tumors with genetic defects in MMR genes have greater susceptibility to immune checkpoint blocking agents(20). Moreover, unaffected family members may be identified and receive genetic counseling and risk assessment to enter pancreatic cancer screening programs and undergo cancer surveillance and prevention (21, 22).
Supplementary Material
Acknowledgments
Financial support: Dr McAllister is a Paul Calabresi K12 clinical scholar (NCI grant awarded to MDACC K12CA088084–16A1) and V Foundation Scholar. Dr McAllister and Dr Maitra have also received support from philanthropic contributions to the University of Texas MD Anderson Pancreatic Cancer Moon Shots Program.
Footnotes
Conflicts of Interest: The authors declare no potential conflicts of interest.
References
- 1.Surveillance, Epidemiology, and End Results Program: SEER Cancer Statistics Factsheets: Pancreas Cancer. Bethesda, MD: National Cancer Institute; 2016. [Available from: http://seer.cancer.gov/statfacts/html/pancreas.html. [Google Scholar]
- 2.Klein AP. Genetic susceptibility to pancreatic cancer. Mol Carcinog. 2012;51(1):14–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brune KA, Lau B, Palmisano E, Canto M, Goggins MG, Hruban RH, et al. Importance of age of onset in pancreatic cancer kindreds. J Natl Cancer Inst. 2010;102(2):119–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kumar S, Torres MP, Kaur S, Rachagani S, Joshi S, Johansson SL, et al. Smoking accelerates pancreatic cancer progression by promoting differentiation of MDSCs and inducing HB-EGF expression in macrophages. Oncogene. 2015;34(16):2052–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Incio J, Liu H, Suboj P, Chin SM, Chen IX, Pinter M, et al. Obesity-Induced Inflammation and Desmoplasia Promote Pancreatic Cancer Progression and Resistance to Chemotherapy. Cancer Discov. 2016;6(8):852–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gullo L, Pezzilli R, Morselli-Labate AM, Italian Pancreatic Cancer Study G. Diabetes and the risk of pancreatic cancer. N Engl J Med. 1994;331(2):81–4. [DOI] [PubMed] [Google Scholar]
- 7.Carreras-Torres R, Johansson M, Gaborieau V, Haycock PC, Wade KH, Relton CL, et al. The Role of Obesity, Type 2 Diabetes, and Metabolic Factors in Pancreatic Cancer: A Mendelian Randomization Study. J Natl Cancer Inst. 2017;109(9). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Murphy KM, Brune KA, Griffin C, Sollenberger JE, Petersen GM, Bansal R, et al. Evaluation of candidate genes MAP2K4, MADH4, ACVR1B, and BRCA2 in familial pancreatic cancer: deleterious BRCA2 mutations in 17%. Cancer Res. 2002;62(13):3789–93. [PubMed] [Google Scholar]
- 9.Su GH, Hruban RH, Bansal RK, Bova GS, Tang DJ, Shekher MC, et al. Germline and somatic mutations of the STK11/LKB1 Peutz-Jeghers gene in pancreatic and biliary cancers. Am J Pathol. 1999;154(6):1835–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lynch HT, Brand RE, Hogg D, Deters CA, Fusaro RM, Lynch JF, et al. Phenotypic variation in eight extended CDKN2A germline mutation familial atypical multiple mole melanoma-pancreatic carcinoma-prone families: the familial atypical mole melanoma-pancreatic carcinoma syndrome. Cancer. 2002;94(1):84–96. [DOI] [PubMed] [Google Scholar]
- 11.Jones S, Hruban RH, Kamiyama M, Borges M, Zhang X, Parsons DW, et al. Exomic sequencing identifies PALB2 as a pancreatic cancer susceptibility gene. Science. 2009;324(5924):217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klein AP, Brune KA, Petersen GM, Goggins M, Tersmette AC, Offerhaus GJ, et al. Prospective risk of pancreatic cancer in familial pancreatic cancer kindreds. Cancer Res. 2004;64(7):2634–8. [DOI] [PubMed] [Google Scholar]
- 13.Klein AP, Hruban RH, Brune KA, Petersen GM, Goggins M. Familial pancreatic cancer. Cancer journal. 2001;7(4):266–73. [PubMed] [Google Scholar]
- 14.Goggins M, Schutte M, Lu J, Moskaluk CA, Weinstein CL, Petersen GM, et al. Germline BRCA2 gene mutations in patients with apparently sporadic pancreatic carcinomas. Cancer Res. 1996;56(23):5360–4. [PubMed] [Google Scholar]
- 15.Shindo K, Yu J, Suenaga M, Fesharakizadeh S, Cho C, Macgregor-Das A, et al. Deleterious Germline Mutations in Patients With Apparently Sporadic Pancreatic Adenocarcinoma. J Clin Oncol. 2017:JCO2017723502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grant RC, Selander I, Connor AA, Selvarajah S, Borgida A, Briollais L, et al. Prevalence of germline mutations in cancer predisposition genes in patients with pancreatic cancer. Gastroenterology. 2015;148(3):556–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holter S, Borgida A, Dodd A, Grant R, Semotiuk K, Hedley D, et al. Germline BRCA Mutations in a Large Clinic-Based Cohort of Patients With Pancreatic Adenocarcinoma. J Clin Oncol. 2015;33(28):3124–9. [DOI] [PubMed] [Google Scholar]
- 18.Hu C, Hart SN, Bamlet WR, Moore RM, Nandakumar K, Eckloff BW, et al. Prevalence of Pathogenic Mutations in Cancer Predisposition Genes among Pancreatic Cancer Patients. Cancer Epidemiol Biomarkers Prev. 2016;25(1):207–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fogelman DR, Wolff RA, Kopetz S, Javle M, Bradley C, Mok I, et al. Evidence for the efficacy of Iniparib, a PARP-1 inhibitor, in BRCA2-associated pancreatic cancer. Anticancer Res. 2011;31(4):1417–20. [PubMed] [Google Scholar]
- 20.Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, Eyring AD, et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N Engl J Med. 2015;372(26):2509–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Canto MI, Harinck F, Hruban RH, Offerhaus GJ, Poley JW, Kamel I, et al. International Cancer of the Pancreas Screening (CAPS) Consortium summit on the management of patients with increased risk for familial pancreatic cancer. Gut. 2013;62(3):339–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McAllister F MM, Uberoi GS, Uberoi AS, Maitra A and Bhutani MS. Current Status and Future Directions for Screening Patients at High Risk for Pancreatic Cancer. Gastroenterology & Hepatology. 2017;In press. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


