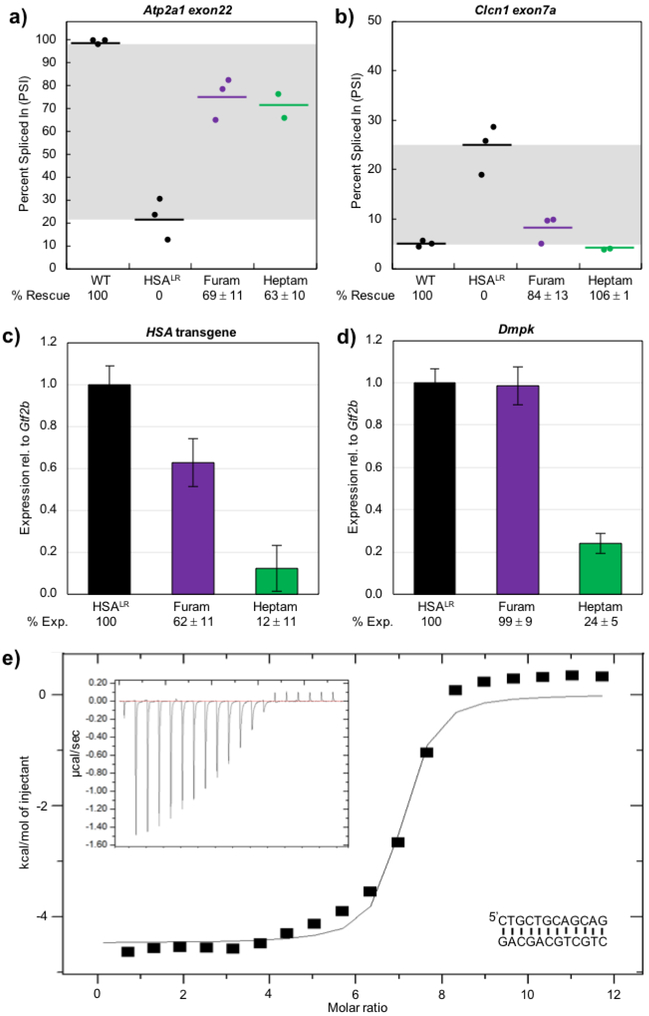
Figure 2. In HSALR mice, furamidine recued Atp2a1 and Clcn1 mis-splicing and reduced HSA transgene levels.
RT-PCR confirmed that both a) Atp2a1 exon22 and b) Clcn1 exon7a mis-splicing events were partially rescued by furamidine (purple) and heptamidine (green) treatment (p<0.01). RT-qPCR data showed that furamidine (purple) reduced c) HSA transgene levels (p<0.01) and did not affect d) endogenous Dmpk levels, while heptamidine (green) treatment drastically reduced both (p<0.001). e) A representative isothermal titration calorimetry (ITC) binding curve using a d(CTGCTGCAGCAG) palindrome sequence (inset lower right) shows that furamidine binds CTG repeat DNA with a K = (2.1 ± 0.3) ×106 M–1 when fit with a single binding model, where N = 7.2 ± 0.6, ∆H = −4104 ± 436 cal/mol, ∆S = 15 ± 1 cal/mol/deg. Raw heat of reaction versus time is inset in the upper left corner.