Abstract
Dynamic modification of cell proteins with phosphate is one of the key regulators of the cellular response to external stimuli. Phosphorylation-based signaling networks mediate cell proliferation, differentiation, and migration, and their dysregulation is the basis of multiple diseases. However, the transient nature of the regulatory protein phosphorylation and low site occupancy mean that only a fraction of the protein is phosphorylated at a given time, and it is a challenge to measure the degree and dynamics of phosphorylation using traditional biochemical means. Technological advances in the field of mass spectrometry (MS) made it possible to generate large sets of phosphoproteomics data, probing the phosphoproteome with great depth, sensitivity, and accuracy. Therefore, quantitative phosphoproteomics emerged as one of the essential components of the systems biology approach for profiling of complex biological networks. Nowadays, the challenge lies in validation of the information and in its integration into the comprehensive models of cell decision processes. This article reviews the role of phosphoproteomics in systems biology, the MS-based approach, and technical details of the methods. Recent examples of quantitative measurements and methodologies as well as applications to the studies of the immune system and infectious diseases are presented and discussed.
INTRODUCTION
The aim of systems biology is to understand the mechanisms ruling the biological systems, and to create dynamic models, which can then be used to predict the possible outcomes of various perturbations of a system. Although usually hypothesis-driven, systems biology relies on collection of large sets of quantitative measurements of changes in the biological components of a system, such as a cell or a population of cells. The components, e.g., transcripts, proteins, protein modifications, and interactions between molecules are dynamic and their spatial and temporal distributions are essential for building reliable and testable models. The recent technological advancements made accurate measurements of many components at once possible, quick, and reproducible.
MS has long been a method of choice for detection of peptides, proteins, and protein modifications in biological preparations. Only recently, however, identification and quantification of many proteins in complex biological mixtures have become possible with the development of experimental and computational methodologies.
Protein phosphorylation is a ubiquitous post-translational modification (PTM), which, due to its reversible and dynamic nature, is crucial in signal transduction, regulating processes like cell proliferation, metabolism, motility, membrane transport, and apoptotic cell death.1 The impact of individual phosphorylation events on cell behavior is immense and deciphering them is key for the systems biology approach to cellular signaling networks.2
Proteins can be reversibly phosphorylated on serine, threonine, and tyrosine residues. The process is enzymatically catalyzed by protein kinases and phosphate groups are removed from proteins by phosphatases. Phosphoproteomics is defined as the study of the complement of proteome that undergoes phosphorylation, the stoichiometry of the protein phosphorylation status at individual residues, and the changes in phosphorylation in response to perturbations. Currently, there are 78,783 vertebrate phosphorylation sites in the PhosphoSite database (http://www.phosphosite.org), 20,002 sites from different eukaryotic species in the PhosphoELM database (http://phospho.elm.eu.org/), and 70,095 sites from eukaryotes and bacteria in the Phosida database (http://www.phosida.org/).
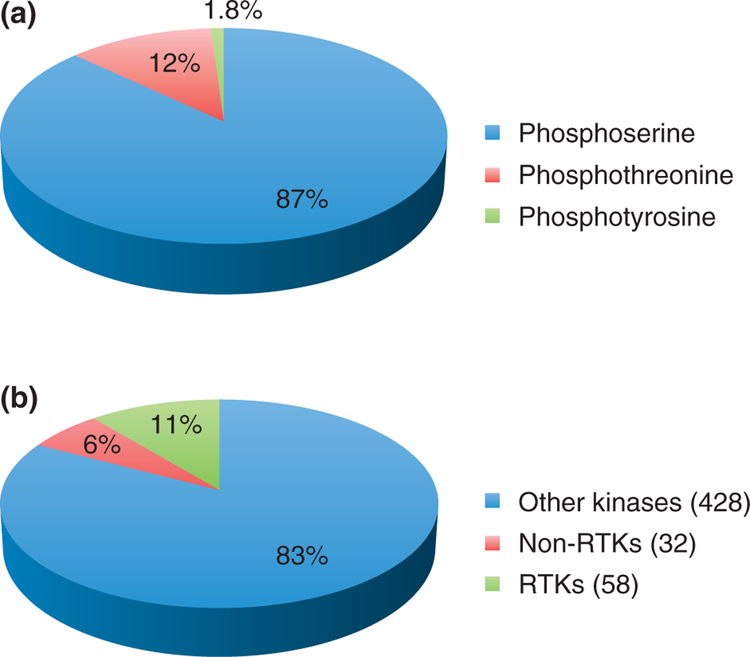
Tyrosine phosphorylation is especially intriguing, because, despite its ultra low abundance [it was initially estimated to constitute 0.05% of the phosphoproteome,3 but recent large-scale studies4 placed the number of tyrosine-phosphorylated peptides in human cells at 1.8%, Figure 1(a)], it is most often involved in disease. Changes in tyrosine phosphorylation signaling networks contribute to many oncogenic malignancies. Many of these changes are caused by dysregulation of kinases or directly by activating and inactivating mutations in kinases and phosphatases.5
FIGURE 1.
(a) Relative abundance of serine, threonine, and tyrosine phosphorylations. (b) Tyrosine kinase family, divided into receptor tyrosine kinase (RTK) and non-receptor tyrosine kinase (non-RTK) subfamilies, in relation to the total number of kinase sequences in the human genome.
Of the 518 kinase sequences encoded by the human genome (1.7%), 430 are expected to be catalytically active.6,7 The tyrosine kinase family, with 90 members [Figure 1(b)], is the largest of 134 families in the human kinome,6 and can be divided into two subfamilies. The RTK subfamily, with 58 members, gathers transmembrane proteins, usually activated by the binding of a ligand to the extracellular region, which causes dimerization and autophosphorylation of the intracellular tyrosine kinase domains. The 32 proteins from the nn-RTK subfamily act rather downstream in the signaling pathways, but can also respond to extracellular stimuli through modification of the modular units such as Src homology 2 (SH2), Src homology 3 (SH3), or pleckstrin homology (PH) domains. Tyrosine phosphorylation temporal dynamics, especially for RTKs (such as epidermal growth factor (EGF) receptor signaling) differs substantially from that of serine/threonine phosphorylation, the former peaking much faster than the latter.4
Phosphorylation plays a pivotal role in the regulation of the immune system function—rapid signaling is essential for changes in adaptive and innate immune system components when dealing with infectious agents, and aberrations result in succumbing to infection or in defects such as allergies and autoimmunity. Tyrosine phosphorylation is key for signal transduction through antigen receptors, integrins, and cytokine receptors. On the other hand, pathogens can induce specific signaling changes in the host to help them evade the immune response. Therefore, identification of phosphorylation sites and their dynamics upon treatment with a plethora of stimulatory and infectious agents is crucial for the development of therapeutic strategies and combating major health challenges of today.
This review highlights quantitative phosphoproteomics by MS, with special focus on the methodology for quantification of tyrosine phosphorylation and recent advances in phosphoproteomic approaches to the studies of the infectious diseases and the immune system.
STRATEGIES FOR MS-BASED PROTEOMIC TYROSINE PHOSPHORYLATION ANALYSIS
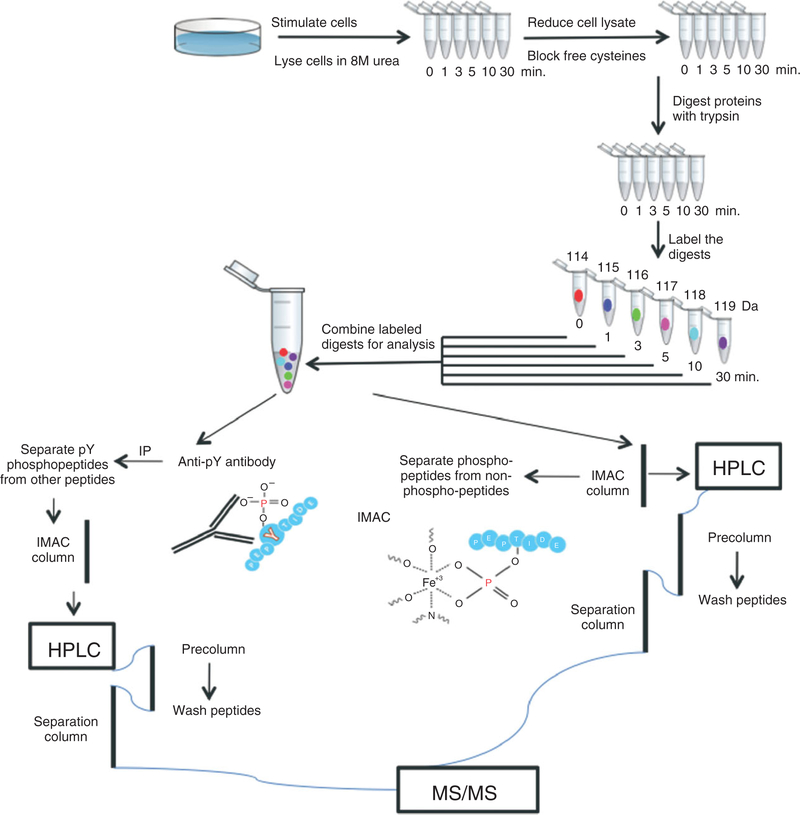
Tyrosine phosphorylation, rare, but indispensable in the cellular regulatory pathways, has been a special challenge in phosphoproteome analysis, because of the dynamic range (from high to extremely low, down to a few copies per cell) of phosphorylated protein levels, transient nature of the modification, heterogeneity of phosphorylation sites on any given protein, and high background of much more abundant serine and threonine phosphorylation sites. Typically, tyrosine phosphorylation sites have been identified on individual proteins, using a combination of protein-specific anti-phosphotyrosine (anti-pTyr) antibodies in Western blot and gel band MS analysis. Immunohistochemistry, immunofluorescence, and flow cytometry are additional methods for probing phosphorylation cascades. In recent years, though, identification and quantification of hundreds of tyrosine phosphorylation sites at a time in complex samples by MS has become a reality. Currently, the workflow (Figure 2) includes a specific pTyr-specific enrichment step in addition to the general enrichment of phosphorylated peptides followed by liquid chromatography directly coupled with ESI-MS.
FIGURE 2.
Example of the workflow for quantitative phosphorylation analysis using iTRAQ labeling. Cultured cells are stimulated with specific ligands for a series of time points and lysed without detergent. The lysates are treated with a reducing agent to break the disulfide bonds and the free cysteine residues are modified to prevent the reformation of disulfide bonds. Subsequently, the lysates are digested with trypsin, the samples are labeled with iTRAQ (up to eight conditions can be distinguished) and combined. For tyrosine phosphorylation analysis, immunoprecipitation (IP) with anti-pTyr antibodies is performed prior to immobilized metal affinity chromatography (IMAC) enrichment; for global phosphorylation analysis, phosphopeptides are enriched only with IMAC. After elution from IMAC column, phosphorylated peptides are analyzed by LC-MS/MS.
Enrichment and Quantification
Anti-pTyr antibodies are used for immunoprecipitation (IP) of tyrosine-phosphorylated proteins and peptides from cell and tissue lysates.8 Commercially available anti-pTyr antibodies can be used to capture the phosphopeptides. The proven specificity of anti-pTyr antibodies is unique, because general anti-phosphoserine (anti-pSer) and anti-phosphothreonine (anti-pThr) antibodies are not available, although there are reports of specific antisera successfully used for subsets of serine- or threonine-phosphorylated peptides.9 Variations of the method include binding of the antibodies to the beads prior to sample application versus incubation of antibody solution with the sample and adding beads as the last step, and IP of whole phosphoproteins directly from cell lysates10,11 versus IP of phosphopeptides from predigested sample.12–15 The experimental optimization is advised for individual experiments, since the differences in sample composition result in suboptimal recovery, when published protocols standardized for another cell type or conditions are used. The antibodies, although pTyr-specific, can display different levels of affinity to surrounding amino acids, so use of the antibody mix and varying the ratios is recommended. The IP supernatant might be checked for remaining pTyr-containing peptides, although they may be very difficult to detect by MS due to low abundance among unbound non-phosphorylated peptides. Only if protein IP is performed, enrichment in tyrosine-phosphorylated peptides can be evaluated by Western blot. Further enrichment of the IP-eluted sample can be performed using affinity chromatography method, such as immobilized metal affinity chromatography (IMAC), or metal oxide affinity chromatography (MOAC), most often used with titanium dioxide (TiO2),16,17 but also with zirconium dioxide,18 both based on the high affinity of acidic peptides in metal ions. IMAC, most often used in phosphoproteomics with metal ions such as Fe(III), Ga(III),19 Zn(II),20 and metal-chelating resins such as POROS-MC, has been widely used offline and online, coupled with nano-LC and ESI-MS. MOAC with use of TiO2 has recently gained popularity and some companies offer mini-columns in pipette tips, a practical alternative to columns with greater bed volume. Since phosphorylated peptides tend to be more acidic than their unmodified counterparts, significant phosphopeptide enrichment can be achieved in this step, although the non-phosphorylated acidic peptides can still be retained, contaminating the preparation. Conversion of peptides to corresponding methyl esters renders IMAC more selective toward phosphorylated peptides and this derivatization method has been successfully used in global phosphorylation analysis.21,22 The IP step in tyrosine phosphorylation analysis, however, can be used in combination with IMAC without methyl esterification and the non-specific binding is negligible.13,23,24 Another commonly used enrichment/fractionation step, strong cation exchange (SCX) utilizes the charge difference between peptides bearing negatively charged phosphate groups and the unmodified peptides for separation.25,26 Although less selective than IMAC and TiO2 enrichments, SCX has been a method of choice for reduction of sample complexity in many studies.
All the enrichment methods described above work well with the labeling approaches used for relative quantification. Labels relying on metabolic incorporation in vivo (e.g., SILAC27) and post-processing chemical derivatization (e.g., iTRAQ28 and iTRAQ 8-plex29) are compatible with anti-pTyr IP and IMAC/MOAC13,23,24 and allow multiplexed analyses within one MS run. If the sample is labeled, and the quantitative changes in tyrosine phosphorylation are to be observed, the sample preparation-related variation has to be excluded. The easy way is to normalize the labeled peptides from the IP supernatant. The peptides from specific proteins unchanged by the experiment, as well as the average ration of supernatant peptides, should be close to 1 and the correction factor can be calculated and applied to the sample. Detailed description of the labeling methods is beyond the scope of this review and they have been reviewed recently,30,31 but it is important to note that growing interest in systems biology use of quantitative MS data has been fueling efforts in bioinformatics-based approaches to label-free and absolute quantification. Some of the recently developed methods for improving the accuracy of absolute quantification and comparisons between many LC-MS runs are not yet applicable to phosphorylated peptides32 but some are being designed with phosphoproteomics in mind,33,34 and offer great promise for the modeling of cellular signaling pathways regulated by phosphorylation. Indeed, these methods may be crucial in moving from analysis of the cells to the whole animal; isotopic labeling, although attempted, is expensive and challenging.35 Other challenges connected with quantitative analysis of whole animals are difficulties in separating different cell populations from tissues and increased errors in data reproducibility.
Instrumentation
Relative stability of pTyr in comparison to serine and threonine phosphorylation facilitates the MS analysis. For tyrosine phosphorylation, there is usually no neutral loss of 80 Da, unlike for serine and threonine-phosphorylated peptides. Instead, the MS/MS spectra of peptides containing pTyr (precursor mass difference +80 Da) are characterized by the presence of a specific pTyr immonium ion of m/z 216.0426, and otherwise produce series of y and b ions, which can be used for peptide sequencing. Consequently, no MS3 (another round of MS, which selectively isolates the backbone peptide after neutral loss in MS/MS, and targets it with higher collision energy, enabling peptide sequencing) or multistage activation (which uses both MS spectra of intact modified peptide and neutral loss MS/MS spectra to get even more complete information about peptide sequence) are necessary to identify the peptide sequence and phosphorylation site.
For tyrosine phosphorylation alone, the stability of the phosphate is not an issue; the quantification method is the most important constraint on the instrument choice. SILAC-labeled samples, quantified on the MS level, yielded good results already on quadrupole time-of-flight (Q-TOF) instruments, but now they are analyzed mostly on instruments equipped with quadrupole ion trap and Fourier- transform-based ion detectors, which seem to provide even more accurate data. iTRAQ-labeled samples, quantified using MS/MS spectra, require instruments capable of registering low m/z ions and with high resolving power, because the marker ions are slightly above 100 Da, a region in the spectra where many contaminants can be found. Numerous successful studies have been performed with hybrid Q-TOF instruments.13–15,23,24 LTQ Orbitrap, which collects collision-induced dissociation (CID) and high-energy collisional dissociation (HCD) spectra sequentially for peptide identification and quantification, respectively, has been shown to gather good quality data.36 Since tyrosine-phosphorylated peptides are often additionally modified on serine and threonine residues (e.g., in human Erk1, residues: T202, Y204 can be phosphorylated and are located in one tryptic peptide24), these labile modifications should be taken into consideration if the study is supposed to be fully informative. Electron transfer dissociation (ETD) has been demonstrated to be especially useful in the detection of labile modifications, such as serine and threonine phosphorylation.37 If it is necessary to choose one mass spectrometer for a variety of phosphoproteomic studies, again, LTQ Orbitrap equipped with ETD seems to be the most versatile instrument.
Careful validation of the data is essential for correct assignment and quantitative analysis of phosphorylation sites. False negative and false positive phosphorylation site assignments occur during automated site assignment. Misassignments in peptides containing more than one potentially phosphorylated amino acid are common. In global phosphorylation, diagnostic neutral loss of 98 Da (phosphoric acid) can be wrongly assigned (especially in low-resolution instruments) and confused with the neutral loss of 2-(methylthio)acetamide, modification introduced during sample preparation using alkylation with iodoacetamide. Other modifications introduced during this process can be mistaken for phosphorylation, e.g., 79 Da mass shift connected with carbamidomethylation with sodium adduction or loss of 80 Da from O-sulfated peptides.38 Manual spectra validation (visual inspection) by an experienced researcher is the best method to distinguish between these artifacts and real peptide phosphorylation, and it is used successfully in tyrosine phosphorylation analyses, with hundreds of sites,12,13 but it is not realistic for the datasets consisting of thousands of phosphorylation sites. For large datasets, the general methods to minimize false identifications of peptides include searches using at least three different algorithms, or using a scrambled database to establish a false positive rate. Commercially available software includes phosphorylation site assignment algorithms (e.g., Ascore in Sequest), and there are constant efforts to develop more effective algorithms (e.g., PhosphoScore,39). Careful validation greatly increases the credibility of the datasets so it is worth the effort even if time consuming.
APPLICATIONS OF PHOSPHOPROTEOMICS IN IMMUNOLOGY
Many combinations of enrichment, quantification, and instrument choices described above have been used to explore the phosphoproteome. In immunology-related studies, the focus so far has been rather on method development than on addressing the systems biology questions. Mainly, global phosphorylation changes in response to stimuli such as cytokines, chemokines, or Toll-like receptor ligands have been explored. There are, however, a handful of reports focusing on tyrosine phosphorylation, especially in the T-cell receptor (TCR) signaling (Table 1). Recently, a few studies of changes in phosphorylation levels upon infection with whole pathogens have been performed and are worth reviewing.
TABLE 1.
Summary of Representative MS-based Tyrosine Phosphoproteomic Studies of the Immune System Signaling
| Cell Type and Stimulation | Enrichment Method | Quantitative? | Number of pTyr Sites Identified/Quantified | Reference |
|---|---|---|---|---|
| Jurkat | Anti-pTyr IP, methyl esterification, IMAC | No | 19 | 40 |
| Jurkat, pervanadate | Anti-pTyr IP, methyl esterification, online IMAC | No | 103 | 10 |
| Jurkat, anti-CD3/CD28 | Anti-pTyr IP, IMAC | Yes (iTRAQ) | 101/87 | 13 |
| Jurkat, pervanadate | Anti-pTyr IP | Yes (dendrimer conjugation) | 75 | 41 |
| Jurkat, Namalwa, anti-CD3, IgM, pervanadate | Anti-pTyr IP | Yes (SILAC) | 299 proteins | 11 |
| Jurkat, anti-CD3, anti-CD4 | Anti-pTyr, IMAC | Yes (SILAC, label-free) | 168 | 42 |
| Primary T-cells, anti-CD3 | Anti-pTyr, IMAC | Yes (iTRAQ) | 77 | 12 |
Anti-pTyr, anti-phosphotyrosine; IP, immunoprecipitation; IMAC, immobilized metal affinity chromatography.
Tyrosine Phosphorylation in TCR Signaling
Activation of T-cells is triggered by co-stimulation of TCR and CD 28 co-receptor, and many proteins involved in the signaling pathway have been identified.43,44 Tyrosine kinases, including Lck and Zap-70, are recruited to the proximity of TCR–CD3 complex, and their phosphorylation results in signaling cascades leading to activation of NF-κB and calcium-dependent pathways. The signaling relies heavily on tyrosine phosphorylation, so the TCR signaling network has been chosen as one of the models for early studies of tyrosine phosphorylation dynamics. Most of the initial phosphoproteomics studies have been conducted with the use of cultured cells (which still are a useful model because of their availability and ease of treatment with ligands or inhibitors), and for TCR signaling studies Jurkat T-cell leukemia cell line has been most widely used. Pioneering study in Jurkat cells stimulated with anti-CD3 and anti-CD4, mimicking TCR ligation, yielded 19 tyrosine-phosphorylated peptides increased after antibody stimulation in an experiment combining enrichment with anti-pTyr IP, methyl esterification and IMAC, followed by LC–ESI–MS.40 Improvements in the technology platform allowed assignment of tyrosine phosphorylation sites on 138 different proteins in Jurkat cells,10 either associated with T-cell-specific functions, or with general cellular processes. With further refinements of the technique and iTRAQ labeling for relative quantification, over 100 tyrosine phosphorylation sites were analyzed in Jurkat cells stimulated with anti-CD3 alone or anti-CD3 and anti-CD28,13 demonstrating differences in tyrosine phosphoproteomes of T-cells receiving only TCR signal or double signal through TCR and co-receptor, which, according to the two-signal theory, is indispensable for T-cell activation.
An alternative method for identification and relative quantification of phosphorylation sites based on dendrimer conjugation coupled with anti-pTyr IP and stable-isotope labeling has been developed by Aebersold’s lab41 and applied to identify and quantify all known tyrosine phosphorylation sites within the immunoreceptor tyrosine-based activation motifs (ITAM) of the TCR–CD3 chains, and previously unknown tyrosine phosphorylation sites on 97 proteins in human T-cells. Jurkat cells activated with pervanadate were used in this study.
A quantitative comparison between T-cell and B-cell signaling has been performed recently.11 Jurkat cells and Namalwa (lymphoblastoid cell line) cells labeled with SILAC were stimulated with anti-CD3 and IgM, respectively, for specific receptor cross-linking stimulation, and with sodium pervanadate, for non-specific tyrosine phosphorylation induction. The comparison, including pervanadate-stimulated non-lymphoid HEK293 cells as a control, uncovered many universal and pathway-specific components of pTyr-mediated signaling.
A comprehensive recent study42 employed multi-disciplinary approaches to couple proteomic discovery and quantification of tyrosine phosphorylation sites in TCR signaling networks in isogenic Zap-70 tyrosine kinase null and reconstituted Jurkat T-cells with elucidation of the biological roles of these sites using site-directed mutagenesis and protein disruption. Here, label-free quantification was used in parallel to SILAC labeling of Jurkat cells.
Cultured cells, although amenable to metabolic labeling and possible to grow in large quantities, are not an ideal model for the studies of the immune system. Also, stimulation with antibodies does not reflect the in vivo T-cell activation, which involves multiple stimuli. Ideally, primary T-cells stimulated with antigen-presenting cells would be necessary to measure the changes upon activation of adaptive immune response to infection. The primary cells are often hard to obtain and the initial amount of protein used in phosphoproteomics experiments on Jurkat cells has been isolated from rather large numbers of cells—from 1 × 10910 to 1 × 107 cells.13 So far, only a couple of phosphoproteomic studies have been extended to the primary T-cells. A global phosphoproteomic analysis of plasma-derived T-cells, utilizing a sequential enrichment strategy with IMAC and TiO2 resulted in identification of 281 phosphorylation sites, but only four of the unambiguously assigned sites were tyrosine phosphorylation sites and no function has been attributed to them.45 The comparison of T-cells isolated from lymph nodes of diabetes-prone (NOD) and resistant (B6.H2g7) mouse strains, stimulated in vitro with anti-CD3 antibodies, and subsequently iTRAQ-labeled, identified 77 tyrosine phosphorylation sites, which could be quantified for comparison between mouse strains and stimulated and unstimulated cells.12
Global Phosphoproteomics
Global phosphorylation-dependent signal transduction has been a focus of many qualitative and semi-quantitative proteomic studies.
One of the interesting early applications of phosphoproteomics to immunological questions was the analysis of the naturally processed phosphopeptides presented by MHC molecules in several EBV-transformed B lymphoblastoid cell lines.46 The detection of phosphopeptides in the pool of more than 10,000 peptides binding class I MHC was facilitated by IMAC enrichment and the phosphorylated peptides were identified in MS using neutral loss scan—only serine and threonine-phosphorylated peptides were detected. Up to 60 phosphorylated peptides could be detected using this method, depending on the cell line.
The phosphoproteomes of other cells of leukocyte lineage such as AML14.2D10 eosinophil cell line47 or WEHI-231 B-cell lymphoma48 upon various treatments have been successfully probed in a qualitative way, identifying novel sites phosphorylated in specific signaling pathways.
A combination of enzymatic (alkaline phosphatase digest following the TiO2 enrichment) and data-mining methods provided a unique approach to profile the phosphoproteome of J774 macrophages upon interferon (IFN)-γ stimulation. A total of 1143 phosphopeptides from 432 different proteins were identified using LTQ Orbitrap and 125 sites exhibited a twofold change with INF-γ exposure.49
Again, examples of global phosphoproteomic analyses in primary immune cells began to appear only recently. Serine and threonine phosphorylations have been assessed in total population of human primary leukocytes derived directly from blood using SCX combined with phosphochip (TiO2) enrichment and high mass resolution Q-TOF instrument, resulting in identification of 960 different phosphorylation sites and in the conclusion that the white blood cell phosphoprotein level is low compared to cancer cells (osteosarcoma U2OS) analyzed in parallel.50 The method for exploring CXCL12-mediated signaling in human chronic lymphocytic leukemia (CLL) cells has been published in 2009.51 Collectively, 1036 phosphopeptides were identified on the total of 251 proteins from all the five time points assessed. The method, consisting of stimulation of primary CLL cells isolated from patient blood, stimulation with recombinant CXCL12, IMAC and LC-MS/MS on a LTQ ion trap, can be applied to alternative studies involving chemokine/receptor signaling networks as well as other types of signaling networks. It is of note that the authors did not label the peptides obtained from different time points, but rather tried to compare the numbers of detected phosphopeptides between different LC-MS/MS analyses, which is a very rough estimation. Therefore, results are semi-quantitative at best, but the study resulted in identification of many novel downstream targets—phosphoproteins with roles in lymphocyte proliferation, differentiation and activation, and immune system development, whose biological roles are being pursued.
Characterization of the changes in phosphoproteome in real disease, i.e., upon challenge of the host with whole pathogens instead of single molecules is a logical step toward characterization of real, complex, and systemic immune responses. Critical host cell receptors interact with pathogen-produced molecules, initiating unique signaling cascades. Pathogen invasion of host cells is generally associated with the activation of NF-kB, mitogen-activated protein kinases (MAPKs) and (IFN) regulatory factor signaling pathways. It is of interest to analyze both host and pathogen signaling pathways during the course of infection (see Box 1: Phosphorylation in pathogenic bacteria). Because experimental design and analysis are obviously much more difficult than in the case of stimulation of cells with purified ligands, the studies are being attempted only just now. An analysis of the phosphoproteome of spleens from mice infected with Bacillus anthracis has been performed recently.52 Phosphopeptides were quantified using a label-free approach and comparison between the results obtained from mice infected with a lethal strain and those infected with an asymptomatic strain yielded 188 significantly altered phosphopeptides, which can translate into presymptomatic diagnostic markers of anthrax infections.
BOX 1. PROTEIN PHOSPHORYLATION IN PATHOGENIC BACTERIA.
Successful pathogens coordinate the action of virulence factors to colonize the host and to evade the subsequent immune response. Many bacteria are able to counteract the host’s immune response. Serine, threonine, and tyrosine phosphorylation have been identified in bacteria. Serine, threonine, and tyrosine kinases are implicated in the control of pathogenicity functions of numerous pathogens, controlling virulence, drug resistance, and immune response evasion by interference with host signaling. The first site-specific phosphoproteomic studies in bacteria yielded long lists of phosphorylated proteins,53,54 but quantitative and time-resolved approaches are needed to truly understand the regulation of pathogen-host interactions.55
CONCLUSION
Technological advances in phosphoproteomics will allow collection of large sets of accurate, time-resolved data suitable for systems biology. Now, we are able to monitor and quantify thousands of phosphorylation sites in different experimental setups. The development of label-free quantification methods and sophisticated instrumentation will facilitate progress in the analysis of rare cell populations in vivo (as demonstrated by the latest proteomic analysis of dendritic cells56). The shift toward absolute quantification will enable the construction of more complex mathematical models with a real predictive power. A steady improvement of all aspects of systems biology and combination of high-throughput genomic, proteomic, single-cell monitoring, and gene knockout data should allow us to closely follow physiological changes in the infection process and immune response and provide a deeper understanding of disease leading to the improvement of clinical applications.
ACKNOWLEDGMENTS
The author thanks members of the Program in Systems Immunology and Infectious Disease Modeling for helpful discussions and Virginie Sjoelund for help with preparation of Figure 2.
REFERENCES
- 1.Seet BT, Dikic I, Zhou MM, Pawson T. Nat Rev Mol Cell Biol 2006, 7:473–483. [DOI] [PubMed] [Google Scholar]
- 2.Hunter T Cell 2000, 100:113–127. [DOI] [PubMed] [Google Scholar]
- 3.Hunter T, Sefton BM. Proc Natl Acad Sci U S A 1980, 77:1311–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, Mortensen P, Mann M. Cell 2006, 127:635–648. [DOI] [PubMed] [Google Scholar]
- 5.Ortutay C, Valiaho J, Stenberg K, Vihinen M. Hum Mutat 2005, 25:435–442. [DOI] [PubMed] [Google Scholar]
- 6.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. Science 2002, 298:1912–1934. [DOI] [PubMed] [Google Scholar]
- 7.Hanks SK. Genome Biol 2003, 4:111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rush J, Moritz A, Lee KA, Guo A, Goss VL, Spek EJ, Zhang H, Zha XM, Polakiewicz RD, Comb MJ. Nat Biotechnol 2005, 23:94–101. [DOI] [PubMed] [Google Scholar]
- 9.Laurence A, Astoul E, Hanrahan S, Totty N, Cantrell D. Eur J Immunol 2004, 34:587–597. [DOI] [PubMed] [Google Scholar]
- 10.Brill LM, Salomon AR, Ficarro SB, Mukherji M, Stettler-Gill M, Peters EC. Anal Chem 2004, 76:2763–2772. [DOI] [PubMed] [Google Scholar]
- 11.Matsumoto M, Oyamada K, Takahashi H, Sato T, Hatakeyama S, Nakayama KI. Proteomics 2009, 9:3549–3563. [DOI] [PubMed] [Google Scholar]
- 12.Iwai LK, Benoist C, Mathis D, White FM. J Proteome Res 2010, 9:3135–3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim JE, White FM. J Immunol 2006, 176:2833–2843. [DOI] [PubMed] [Google Scholar]
- 14.Moser K, White FM. J Proteome Res 2006, 5:98–104. [DOI] [PubMed] [Google Scholar]
- 15.Zhang Y, Wolf-Yadlin A, Ross PL, Pappin DJ, Rush J, Lauffenburger DA, White FM. Mol Cell Proteomics 2005, 4:1240–1250. [DOI] [PubMed] [Google Scholar]
- 16.Klemm C, Otto S, Wolf C, Haseloff RF, Beyermann M, Krause E. J Mass Spectrom 2006, 41:1623–1632. [DOI] [PubMed] [Google Scholar]
- 17.Larsen MR, Thingholm TE, Jensen ON, Roepstorff P, Jorgensen TJ. Mol Cell Proteomics 2005, 4:873–886. [DOI] [PubMed] [Google Scholar]
- 18.Feng S, Ye M, Zhou H, Jiang X, Zou H, Gong B. Mol Cell Proteomics 2007, 6:1656–1665. [DOI] [PubMed] [Google Scholar]
- 19.Sykora C, Hoffmann R, Hoffmann P. Protein Pept Lett 2007, 14:489–496. [DOI] [PubMed] [Google Scholar]
- 20.Kinoshita E, Yamada A, Takeda H, Kinoshita-Kikuta E, Koike T. J Sep Sci 2005, 28:155–162. [DOI] [PubMed] [Google Scholar]
- 21.Ficarro SB, McCleland ML, Stukenberg PT, Burke DJ, Ross MM, Shabanowitz J, Hunt DF, White FM. Nat Biotechnol 2002, 20:301–305. [DOI] [PubMed] [Google Scholar]
- 22.Ficarro SB, Salomon AR, Brill LM, Mason DE, Stettler-Gill M, Brock A, Peters EC. Rapid Commun Mass Spectrom 2005, 19:57–71. [DOI] [PubMed] [Google Scholar]
- 23.Schmelzle K, Kane S, Gridley S, Lienhard GE, White FM. Diabetes 2006, 55:2171–2179. [DOI] [PubMed] [Google Scholar]
- 24.Wolf-Yadlin A, Kumar N, Zhang Y, Hautaniemi S, Zaman M, Kim HD, Grantcharova V, Lauffenburger DA, White FM. Mol Syst Biol 2006, 2:54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ballif BA, Villen J, Beausoleil SA, Schwartz D, Gygi SP. Mol Cell Proteomics 2004, 3:1093–1101. [DOI] [PubMed] [Google Scholar]
- 26.Beausoleil SA, Jedrychowski M, Schwartz D, Elias JE, Villen J, Li J, Cohn MA, Cantley LC, Gygi SP. Proc Natl Acad Sci U S A 2004, 101:12130–12135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, Mann M. Mol Cell Proteomics 2002, 1:376–386. [DOI] [PubMed] [Google Scholar]
- 28.Ross PL, Huang YN, Marchese JN, Williamson B, Parker K, Hattan S, Khainovski N, Pillai S, Dey S, Daniels S, et al. Mol Cell Proteomics 2004, 3:1154–1169. [DOI] [PubMed] [Google Scholar]
- 29.Choe L, D’Ascenzo M, Relkin NR, Pappin D, Ross P, Williamson B, Guertin S, Pribil P, Lee KH. Proteomics 2007, 7:3651–3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tedford NC, Hall AB, Graham JR, Murphy CE, Gordon NF, Radding JA. Proteomics 2009, 9:1469–1487. [DOI] [PubMed] [Google Scholar]
- 31.Nita-Lazar A, Saito-Benz H, White FM. Proteomics 2008, 8:4433–4443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vogel C, Marcotte EM. Nat Biotechnol 2009, 27:825–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Picotti P, Rinner O, Stallmach R, Dautel F, Farrah T, Domon B, Wenschuh H, Aebersold R. Nat Methods 2010, 7:43–46. [DOI] [PubMed] [Google Scholar]
- 34.Picotti P, Bodenmiller B, Mueller LN, Domon B, Aebersold R. Cell 2009, 138:795–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kruger M, Moser M, Ussar S, Thievessen I, Luber CA, Forner F, Schmidt S, Zanivan S, Fassler R, Mann M. Cell 2008, 134:353–364. [DOI] [PubMed] [Google Scholar]
- 36.Olsen JV, Macek B, Lange O, Makarov A, Horning S, Mann M. Nat Methods 2007, 4:709–712. [DOI] [PubMed] [Google Scholar]
- 37.Syka JE, Coon JJ, Schroeder MJ, Shabanowitz J, Hunt DF. Proc Natl Acad Sci U S A 2004, 101:9528–9533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kruger R, Hung CW, Edelson-Averbukh M, Lehmann WD. Rapid Commun Mass Spectrom 2005, 19:1709–1716. [DOI] [PubMed] [Google Scholar]
- 39.Ruttenberg BE, Pisitkun T, Knepper MA, Hoffert JD. J Proteome Res 2008, 7:3054–3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Salomon AR, Ficarro SB, Brill LM, Brinker A, Phung QT, Ericson C, Sauer K, Brock A, Horn DM, Schultz PG, et al. Proc Natl Acad Sci U S A 2003, 100:443–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tao WA, Wollscheid B, O’Brien R, Eng JK, Li XJ, Bodenmiller B, Watts JD, Hood L, Aebersold R. Nat Methods 2005, 2:591–598. [DOI] [PubMed] [Google Scholar]
- 42.Nguyen V, Cao L, Lin JT, Hung N, Ritz A, Yu K, Jianu R, Ulin SP, Raphael BJ, Laidlaw DH, et al. Mol Cell Proteomics 2009, 8:2418–2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kane LP, Lin J, Weiss A. Curr Opin Immunol 2000, 12:242–249. [DOI] [PubMed] [Google Scholar]
- 44.Lin J, Weiss A. J Cell Sci 2001, 114:243–244. [DOI] [PubMed] [Google Scholar]
- 45.Carrascal M, Ovelleiro D, Casas V, Gay M, Abian J. J Proteome Res 2008, 7:5167–5176. [DOI] [PubMed] [Google Scholar]
- 46.Zarling AL, Ficarro SB, White FM, Shabanowitz J, Hunt DF, Engelhard VH. J Exp Med 2000, 192:1755–1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ryu SI, Kim WK, Cho HJ, Lee PY, Jung H, Yoon TS, Moon JH, Kang S, Poo H, Bae KH, et al. J Biochem Mol Biol 2007, 40:765–772. [DOI] [PubMed] [Google Scholar]
- 48.Shu H, Chen S, Bi Q, Mumby M, Brekken DL. Mol Cell Proteomics 2004, 3:279–286. [DOI] [PubMed] [Google Scholar]
- 49.Marcantonio M, Trost M, Courcelles M, Desjardins M, Thibault P. Mol Cell Proteomics 2008, 7:645–660. [DOI] [PubMed] [Google Scholar]
- 50.Raijmakers R, Kraiczek K, de Jong AP, Mohammed S, Heck AJ. Anal Chem 2010, 82:824–832. [DOI] [PubMed] [Google Scholar]
- 51.O’Hayre M, Salanga CL, Dorrestein PC, Handel TM. Methods Enzymol 2009, 460:331–346. [DOI] [PubMed] [Google Scholar]
- 52.Manes NP, Dong L, Zhou W, Du X, Reghu N, Kool AC, Choi D, Bailey CL, Petricoin EF III, Liotta LA, et al. Mol Cell proteomics 2010. In press. [DOI] [PMC free article] [PubMed]
- 53.Macek B, Gnad F, Soufi B, Kumar C, Olsen JV, Mijakovic I, Mann M. Mol Cell Proteomics 2008, 7:299–307. [DOI] [PubMed] [Google Scholar]
- 54.Macek B, Mijakovic I, Olsen JV, Gnad F, Kumar C, Jensen PR, Mann M. Mol Cell Proteomics 2007, 6:697–707. [DOI] [PubMed] [Google Scholar]
- 55.Jers C, Soufi B, Grangeasse C, Deutscher J, Mijakovic I. Expert Rev Proteomics 2008, 5:619–627. [DOI] [PubMed] [Google Scholar]
- 56.Luber CA, Cox J, Lauterbach H, Fancke B, Selbach M, Tschopp J, Akira S, Wiegand M, Hochrein H, O’Keeffe M, et al. Immunity 2010, 32:279–289. [DOI] [PubMed] [Google Scholar]