In a recent issue of Leukemia, Dong et al. reported the crystal structure of an isolated second homeodomain of human DUX4 (DUX4HD2) bound to a double-stranded DNA fragment (strand-1: 5’-TTCTAATCTAATCX-3’, X=A according to Materials and methods of the original paper[1], annealed with strand-2: 5’-AAGATTAGATTAGT-3’, whereas the sequence record of PDB entry 5z2t shows X=TT. We also note that 5’-TGATTAGATTAGAA-3’ in Materials and methods of the original paper is the reverse sequence of ‘strand-2’ above, which is from the PDB.) containing the sequence that authors refer to as the DUX4 responsive element (DRE), 5’-TAATCTAAT-3’ [1]. The reported structure consisted of two molecules of DUX4HD2 bound to DNA in a head-to-tail arrangement, with each DUX4HD2 recognizing a TAAT motif on the same strand. We were surprised by this configuration, because the DUX4 recognition sequence as defined by ChIP-seq[2, 3] and by mutagenesis[4] is actually 11-nucleotide (nt) long, not 9 nt, namely, 5’-TAATCTAATCA-3’. The terminal CA nucleotides are among the most highly conserved, but an explanation for their necessity is not provided by the structure presented. An additional curiosity was the fact that the apparently critical TAAT sequences are not present in the mouse Dux consensus sequence, which nevertheless has many residues in common with that of human DUX4, and also includes a strongly conserved CA at positions 10 and 11: 5’-TGATTCAATCA-3’ [5, 6].
The authors discussed that the side chain of Arg148 makes a key sequence-specific contact, forming hydrogen bonds with N6 and N7 positions of an adenine base of the TAAT motif in the major groove of DNA (Fig. 2f in the original paper)[1]. The model refined to 2.6 Å resolution (PDB ID: 5z2t) indeed shows distance between the nitrogen atoms from adenine N6 amino group and Arg148 guanidinium group of 2.9 and 3.0 Å for the two DUX4HD2 molecules. However, we note that this is not a favorable interaction in physiological conditions, as both these groups would have positive electrostatic potential and are not compatible with hydrogen-bonding. These unexpected structural features prompted us to re-examine the authors’ crystallographic data available in the protein data bank (PDB: www.rcsb.org)[7]. Examination of the electron density map calculated from the deposited structure factors and atomic coordinates using the program PHENIX[8] suggested that DNA had been misplaced with an incorrect register, as evidenced by strong mFo-DFc difference densities near either end of the DNA molecule (Fig. 1a, c). We have re-modeled and refined the structure with a correctly placed DNA molecule (Fig. 1b, d), which better accounts for the electron density and improved Rwork/Rfree from 27.6/29.9 to 24.6/27.9 % (Supplementary Table 1). In this revised model, the two DUX4HD2 molecules both recognize an AGAT motif fortuitously present tandemly in the DNA substrate used (5’-AGATTAGAT-3’) (Fig. 2). Arg148 side chain forms hydrogen-bonds with the O6 and N7 atoms of a guanine base, which makes better chemical sense than the Arg-adenine interaction in the original model. We have deposited our re-refined structure in the PDB under the accession code 6dfy. Based on these observations, we conclude that the interaction of DUX4HD2 with 5’-TAATCTAAT-3’ sequence is not supported by the authors’ structural data. Because the interactions with AGAT do not fit into the known DRE, and furthermore because HD1 is not accounted for in this study, we advise caution in inferring that the head-to-tail structure adopted by two units of the isolated second DUX4 homeodomain (HD2) in this crystal is representative of the structure formed by the double homeodomains (HD1-HD2) of DUX4 in vivo, or by DUX4 fusions associated with B-cell leukemia.
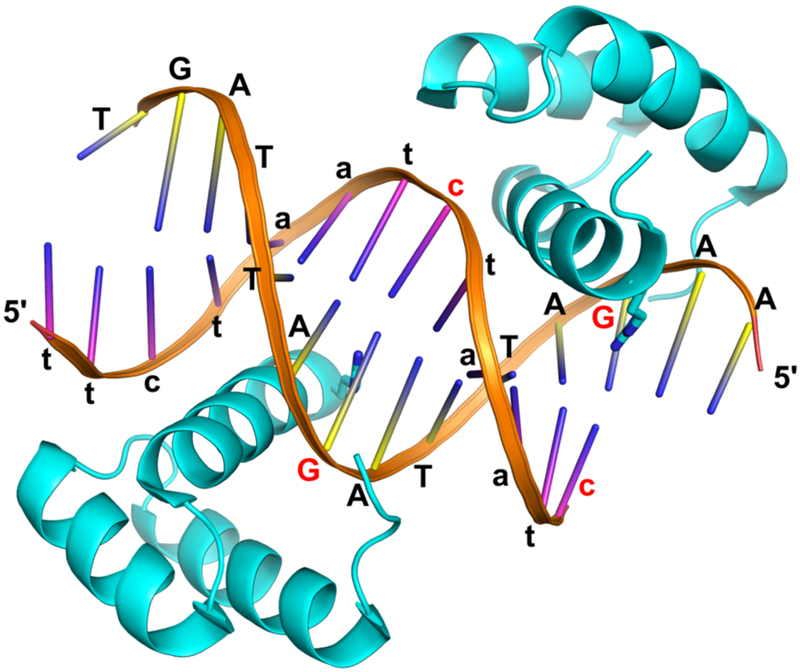
Figure 2.
Re-refined structure of the DUX4HD2-DNA complex. The ‘AA’ dinucleotide at the 5’ end of strand-2 (yellow ladder, capital letters) base-pairs with ‘tt’ at the 5’ end of strand-1 (pink, lower case) from a neighboring complex, facilitating the formation of a pseudo-continuous double helix in the crystal lattice. This crystal contact involves two inter-molecular Hoogsteen A-T base-pairs, as shown in Fig. 1b. The 3’-terminal nucleotide(s) ‘X’ of strand-1 is displaced and disordered. The two guanine bases hydrogen-bonded to Arg148 and their complementary cytosines are highlighted in red.
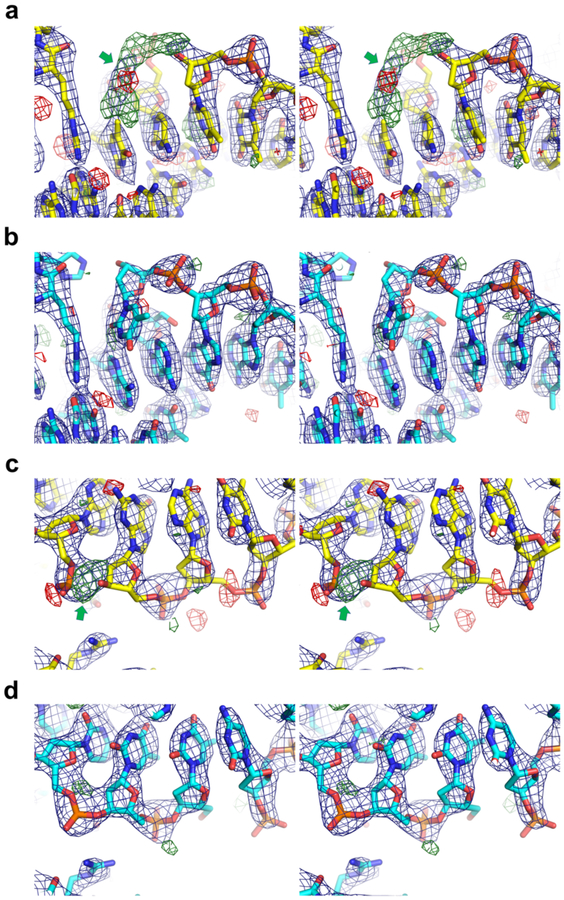
Figure 1.
2mFo-DFc (blue, contoured at 1.5σ) and mFo-DFc (green/red, 3.0σ/–3.0σ) electron densities for the original (a and c) and re-refined (b and d) structures for two representative regions, shown in wall-eye stereo views. The original and re-refined models are shown in yellow and cyan sticks, respectively. The red cross-hairs in a represent water molecules. Strong positive difference density corresponding to the missing nucleotide (a) or phosphodiester bond (c) is indicated by the green arrows.
Supplementary Material
Footnotes
Conflict of Interest:
The authors declare that they have no conflict of interest.
Reference:
- 1.Dong X, Zhang W, Wu H, Huang J, Zhang M, Wang P, et al. Structural basis of DUX4/IGH-driven transactivation. Leukemia. 2018. Epub 2018/03/25. doi: 10.1038/s41375-018-0093-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Geng LN, Yao Z, Snider L, Fong AP, Cech JN, Young JM, et al. DUX4 activates germline genes, retroelements, and immune mediators: implications for facioscapulohumeral dystrophy. Dev Cell. 2012;22(1):38–51. Epub 2012/01/03. doi: 10.1016/j.devcel.2011.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eidahl JO, Giesige CR, Domire JS, Wallace LM, Fowler AM, Guckes SM, et al. Mouse Dux is myotoxic and shares partial functional homology with its human paralog DUX4. Hum Mol Genet. 2016;25(20):4577–89. doi: 10.1093/hmg/ddw287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang Y, Lee JK, Toso EA, Lee JS, Choi SH, Slattery M, et al. DNA-binding sequence specificity of DUX4. Skelet Muscle. 2016;6:8 Epub 2016/01/30. doi: 10.1186/s13395-016-0080-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Whiddon JL, Langford AT, Wong CJ, Zhong JW, Tapscott SJ. Conservation and innovation in the DUX4-family gene network. Nat Genet. 2017;49(6):935–40. Epub 2017/05/02. doi: 10.1038/ng.3846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hendrickson PG, Dorais JA, Grow EJ, Whiddon JL, Lim JW, Wike CL, et al. Conserved roles of mouse DUX and human DUX4 in activating cleavage-stage genes and MERVL/HERVL retrotransposons. Nat Genet. 2017;49(6):925–34. Epub 2017/05/02. doi: 10.1038/ng.3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, et al. The Protein Data Bank. Nucleic Acids Res. 2000;28(1):235–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):213–21. Epub 2010/02/04. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.