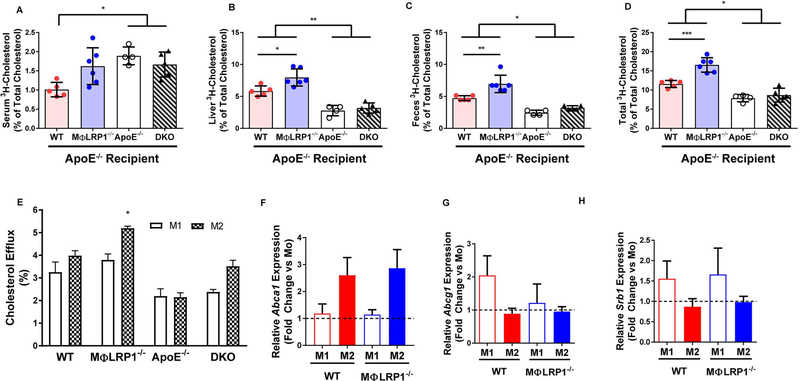
Figure 4. In Vivo and In Vitro Reverse Cholesterol Transport (RCT).
Peritoneal macrophages (MPMs) from wildtype (WT) and MΦLR1−/−, apoE−/−, and apoE−/−/MΦLR1−/−(DKO) mice were cultured with acetylated LDL (50 μg/mL) containing 3H-cholesterol for 48 hours. After equilibration, macrophages were injected into mice of their respective donor genotype undergoing regression (chow diet). RCT was determined as described in Materials and Methods. 3H-cholesterol in (A) serum, (B)liver, (C) feces, and (D) total of all three compartments (% of total cholesterol).(E) In vitro cholesterol efflux to ApoAl in Ml and M2 MPMs from WT and MΦLR1−/−, apoE−/− and DKO mice was determined as described in Materials and Methods. WT and MΦLRP1−/− MPMs were first primed (Mo) and then differentiated to Ml or M2 polarizations as described in Materials and Methods. qRT-PCR was used to determine changes in gene expression of (F) A bcal,(G) Abcgl and (H) Srbl and reported as fold change over IFNγ primed macrophages (Mo; black dashed line). Gene expression data are presented as mean ± SEM. For RCT one-way ANOVA with Bonferroni’s post hoc tests were used to compare effects across genotypes. For cholesterol efflux and gene expression, two-way ANOVA with Sidak’s multiple comparisons test was used to compare effects across genotypes and macrophage subtypes. ***P<0.001, **P<0.01, *P<0.05.