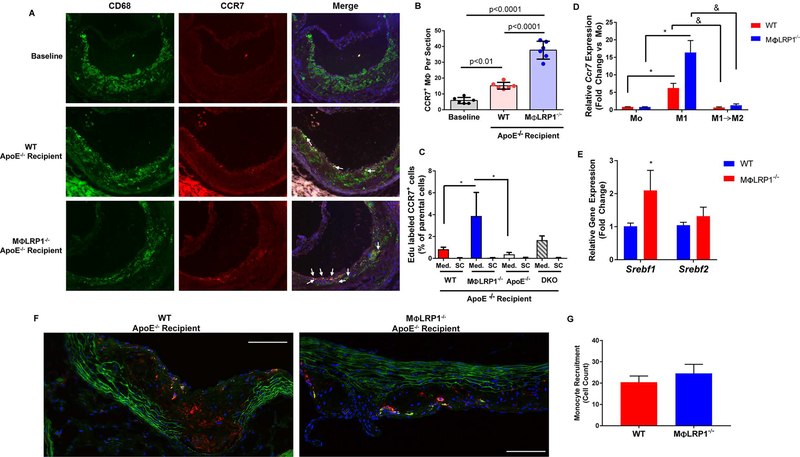
Figure 6. CCR7-dependent Macrophage Egress from Plaques During Regression.
(A) Immunofluorescent staining of CD68 (green), CCR7 (red) and Hoechst (blue) in plaques from baseline and wildtype (WT) and MΦLR1−/− recipient mice undergoing regression. Scale bar = 100μm. (B) Percentage of CD68+ CCR7+ double positive macrophages (white arrows) relative to total CD68+ macrophage numbers. One-way ANOVA with Bonferroni’s post hoc test was used to compare effects among genotypes. (C) EDU-labeled CD11b monocytes from WT, MΦLR1−/−, apoE−/− and DKO donor mice were injected into recipient mice of their respective genotype on chow diet (n=4–6 mice/genotype). After 72 hours mice were sacrificed and in vivo macrophage egress into the mediastinal lymph nodes (Med.) and superficial cervical control lymph nodes (SC) was determined via FACS analysis of EDU+ CCR7+ double-positive cells as described in Materials and Methods. ANCOVA was performed to determine differences in Med. lymph nodes using donor genotype as the independent variable and paired SC lymph nodes as the covariate. Tukey’s post hoc test was used to determine pairwise differences in Med. lymph nodes. *P<0.05. (D) qRT-PCR was used to determine Ccr7 expression in mouse peritoneal macrophages (n=5–6/genotype) primed with IFNγ (50ng/mL; Mo) (dark red and dark blue bars) or differentiated to M1 (open bars) or switched from M1 to M2 (pink and light blue bars) as described in Materials and Methods. Data are presented as mean ± SEM and Wilcoxon Signed Rank test was performed to determine differences from Mo (*P<0.05). Unpaired Mann-Whitney test was performed to determined individual differences between groups (&P<0.05). (E) qRT-PCR was used to determine changes in Srebf1 and Srebf2 expression in MPMs isolated from WT and MΦLR1−/−mice (n=6 and n=5 respectively) and differentiated to the M1 phenotype. Data are represented as mean ± SEM and normalized to the WT recipient group. Unpaired Mann Whitney test was used to compare differences in gene expression between genotypes. *P<0.05. CD11b monocytes isolated from WT and MΦLR1−/− donor mice were labeled with CMFDA Green Cell Tracker and injected i.v. into WT and MΦLRP1−/− recipient mice (n=3 and n=4 respectively) undergoing regression (2×106 monocytes/mouse). After 72 hours, mice were sacrificed and aortic sinus tissue was sectioned. (F) Aortic sinus sections were immunofluorescently stained for CD68 (red) and DAPI (blue) as described in Materials and Methods. Scale bar = 100μm. (G) Monocyte recruitment was determined by counting newly recruited monocyte/macrophages (green) positive for CD68 and normalized to lesion area. Student’s t-test was performed to determine differences in monocyte recruitment.