Figure 1.
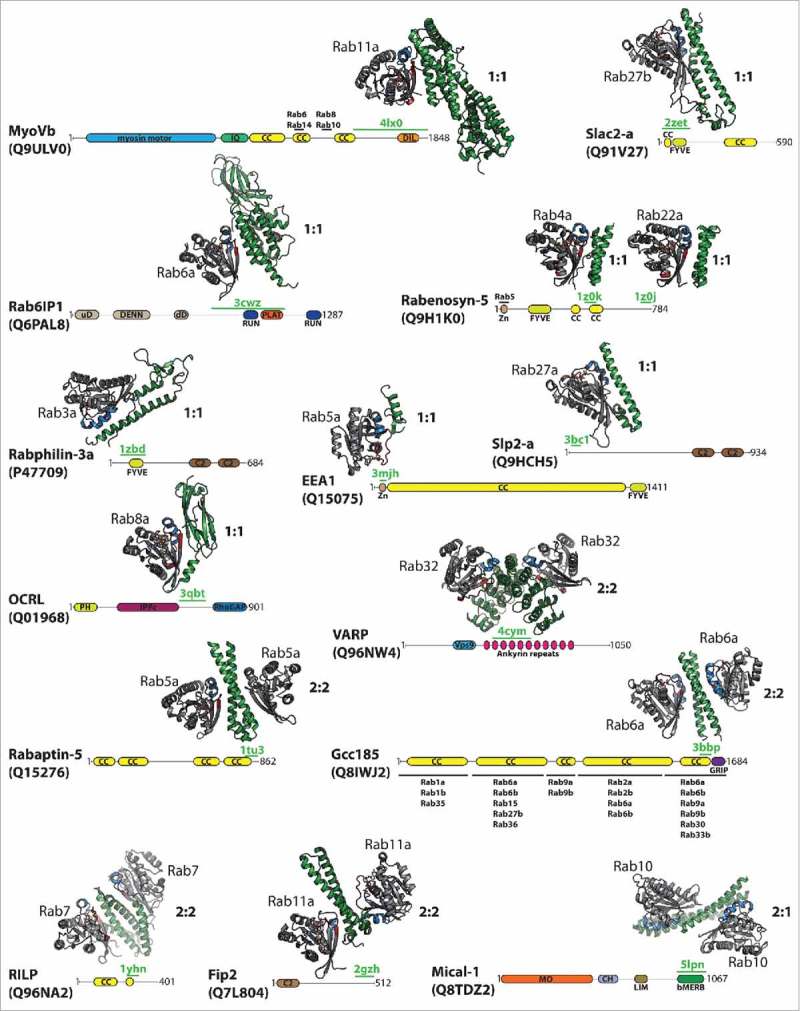
Structurally characterized Rab:effector complexes. The structures of several Rab:effector complexes are indicated together with a scheme showing the overall domain architecture of the effector proteins (according to the SMART and uniprot databases42,43). The structurally resolved regions involved in Rab-binding are highlighted by a green line including the corresponding pdb code, additional Rab-binding sites/regions are indicated by a black line including the Rab proteins known to bind at the corresponding site (the additional Rab binding sites in MyoVb are inferred from studies on MyoVa37). Structures of the Rab:effector complexes are shown in cartoon style (Rab: gray with switch I in red and switch II in blue; effector: green). Besides the Rab-binding regions, Rab effectors usually contain several additional domains, as indicated (bMERB – bivalent Mical/EHBP Rab-binding domain; C2 – C2 Ca2+-binding domain; CC – (predicted) coiled-coil regions; CH – calponin-homology domain; DENN – Rab GEF domain (uD – upstream DENN, dD – downstream DENN); DIL – Dilute domain; FYVE – FYVE Zinc finger domain; GRIP - golgin-97, RanBP2α, Imh1p and p230/golgin-245 (Golgi targeting) domain; IPPc - Inositol polyphosphate phosphatase; IQ – IQ Calmodulin-binding motif; LIM – Zinc-binding domain (Lin-11, Isl-1, Mec-3); MO – Monooxygenase domain; PLAT – Polycystin-1, Lipoxygenase, α-Toxin domain; RUN – RUN domain; Vps9 – Vps9 GEF domain; Zn – Zinc finger C2H2 domain).
