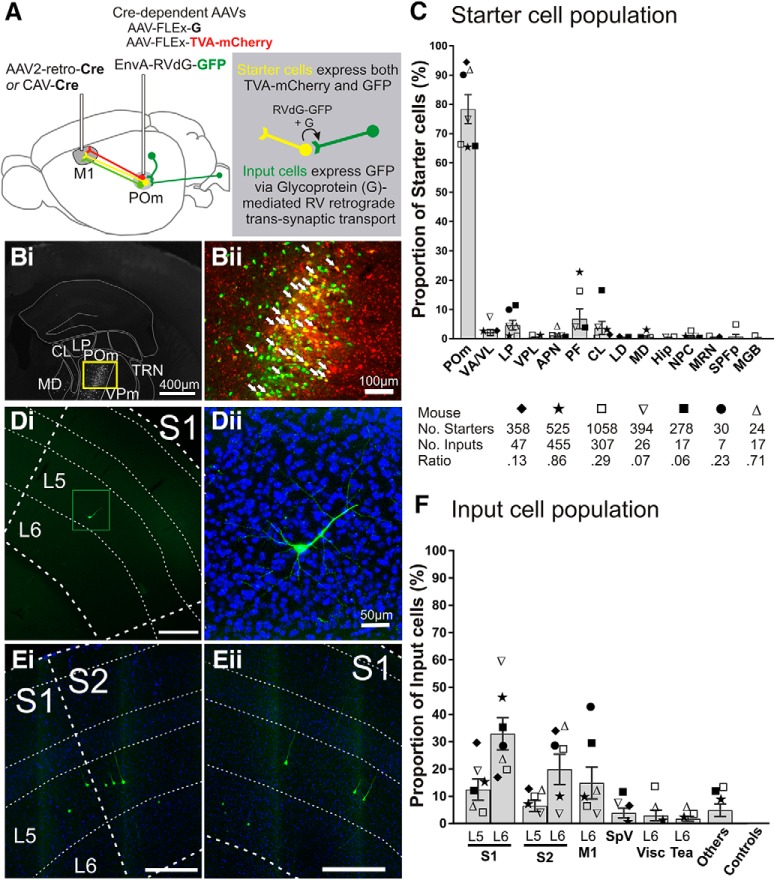
Figure 1.
Trans-synaptic tracing of the inputs to the POm → M1 pathway. A, Schematic of pseudotyped rabies virus-based tracing to label monosynaptic inputs to POm cells projecting to M1. CAV-Cre or AAV2-retro-Cre injected into M1 retrogradely transports to POm where two Cre-dependent AAVs are injected, AAV-CAG-FLExLoxP-TVA-mCherry and AAV-CAG-FLExLoxP-G. Therefore, M1-projecting POm cells are the only cells that can express the TVA receptor, which is recognized by the ligand EnvA, and glycoprotein (G), the envelope protein required retrograde trans-transport of rabies virus. Two weeks later, EnvA-pseudotyped, G-deleted rabies virus tagged with GFP (EnvA-RVdG-GFP) is injected into POm (green projections). Starter cells (yellow projections) are those expressing both TVA-mCherry and GFP. After an additional 4–8 d, starter cells that also express glycoprotein G allow the RVdG-GFP to bud out of the cell and retrogradely spread across the synapse resulting in GFP expressed in the input cells (dark green projections). Therefore, input cells make direct presynaptic contact with M1-projecting POm starter cells. Bi, Representative coronal epifluorescence image of RVdG-infected GFP-expressing cells in POm. Bii, Higher magnification of yellow box in Bi showing starter cells expressing both GFP and TVA-mCherry (overlay). White arrows indicate a subset of starter cells. C, Starter cell locations as a proportion of total starter cell population for each mouse. The majority of starter cells were located in POm (78.4 ± 5.0%), but were also found in nuclei surrounding POm, which also project to frontal and motor areas. Each symbol represents a mouse (n = 7) and the total numbers of starter cells, input cells, and ratios are noted below. D, E, Representative epifluorescence images of GFP-labeled input cells in S1 and S2. S1 layer 5 cell from mouse ▿ in Di is magnified (green box) with DAPI stain in Dii. Cells in Ei and ii are from mouse ★. Scale bars are 300 μm unless otherwise noted. F, Input cell locations as a proportion of total inputs for each mouse. Symbols correspond to those in C. Error bars indicate SEM. APN, Anterior pretectal nucleus; CL, central lateral nucleus; Hip, hippocampus; LD, lateral dorsal nucleus; MD, mediodorsal nucleus; MGB, medial geniculate nucleus, including ventral, medial, and dorsal regions; MRN, midbrain reticular nucleus; NPC, nucleus of the posterior commissure; Tea, temporal association areas; Visc, visceral area; VPL, ventral posterolateral nucleus.